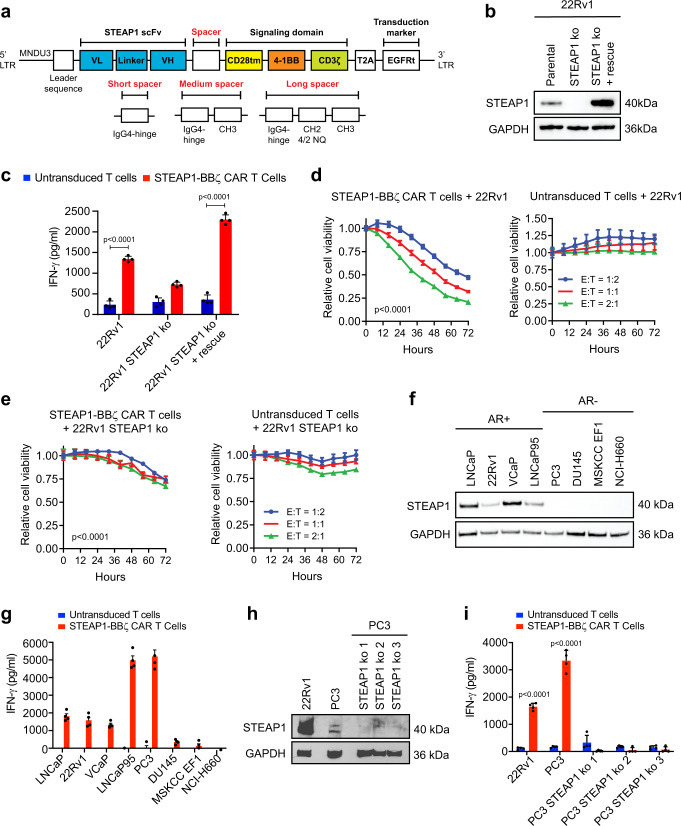
Fig. 2. Screening second-generation 4-1BB chimeric antigen receptors (CARs) to identify a lead for STEAP1 CAR T cell therapy.
a Schematic of the lentiviral STEAP1 CAR construct and variation based on short, medium, and long spacers. LTR long terminal repeat, MNDU3 Moloney murine leukemia virus U3 region, scFv single-chain variable fragment, VL variable light chain, VH variable heavy chain, tm transmembrane, EGFRt truncated epidermal growth factor receptor, 4/2 NQ = CH2 domain mutations to prevent binding to Fc-gamma receptors. b Immunoblots of STEAP1 in 22Rv1 parental cells, STEAP1 knockout (ko) cells, and STEAP1 ko cells with rescue of STEAP1. c IFN-γ enzyme-linked immunosorbent assay (ELISA) results from co-cultures of either untransduced T cells or STEAP1-BBζ CAR T cells with each of the 22Rv1 sublines at a 1:1 ratio at 24 h (p < 0.001). Relative cell viability of (d) 22Rv1 and (e) 22Rv1 STEAP1 ko target cells over time measured by fluorescence live cell imaging upon co-culture with (left) STEAP1-BBζ CAR T cells (p < 0.001) or (right) untransduced T cells at variable effector-to-target (E:T) cell ratios. f Immunoblots demonstrating expression of STEAP1 in androgen receptor (AR)-positive human prostate cancer cell lines but not AR-negative prostate cancer cell lines. g IFN-γ quantification by ELISA from co-cultures of either untransduced T cells or STEAP1-BBζ CAR T cells with each of the human prostate cancer cell lines in (f) at a 1:1 ratio at 24 h. h Immunoblots for STEAP1 in 22Rv1, PC3, and PC3 STEAP1 ko sublines. i IFN-γ quantification by ELISA from co-cultures of either untransduced T cells or STEAP1-BBζ CAR T cells with each cell line in (h) at a 1:1 ratio at 24 h (p < 0.001). For panels (c–e, g and i) n = 4 biological replicates per conditions were used and error bars represent mean with SEM. Panel (b, f, h) displays results representative of n = 3 biological replicates. GAPDH was used as a protein loading control. For panel (c) and (i), two-way ANOVA with Sidak’s multiple comparison test was used. For panels (d) and (e), two-way ANOVA with Tukey’s multiple comparisons test was used. Source data are provided in the Source Data file.