Figure 3.
Knocked in hF9 produced hFIX in hepatocytes
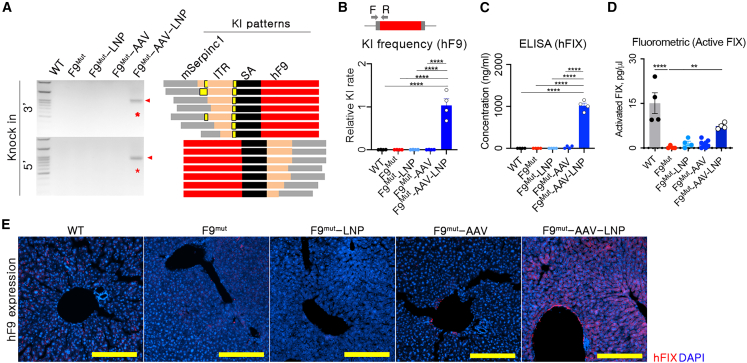
(A) PCR and Sanger sequencing-based genotyping for analyzing KI and its integration pattern. Primers were designed for both sides of the KI-expected regions. Sequencing was conducted using 10 randomly selected clones from each KI-positive amplicon of F9Mut-AAV-LNP (red bar: KI sequence, mSerpinc1: endogenous sequence of Serpinc1; ITR, inverted terminal repeat; SA, splicing acceptor; yellow bar: unexpected indel). (B) Relative KI rates, as analyzed using quantitative PCR in the 3′ site of the expected KI locus. The KI rate was normalized to that of the F9Mut-AAV-LNP group. (C) hFIX concentration in the blood, measured using ELISA (n = 4 per group). Each dot represents data from an individual mouse. Data are presented as mean ± standard error of mean, calculated using one-way ANOVA. (∗∗p < 0.01 and ∗∗∗∗p < 0.0001. (D) Active hFIX in the blood was measured using a FIX activity assay kit (Fluorometric) (n = 4 per group). (E) Immunofluorescence images showing liver hFIX expression (100×). Yellow scale bar: 200 μm.