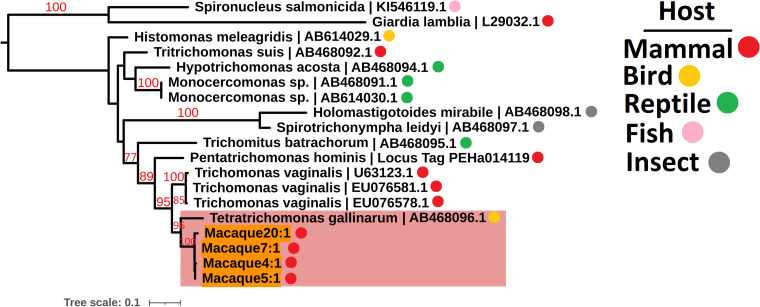
Fig. 2.
Maximum likelihood phylogeny (TIM2e model with equal base frequencies and the discrete gamma model) of Parabasalia-like actin sequences from the macaque fecal metatranscriptome alongside a range of Parabasalia species. Phylogeny is rooted using sequences from Giardia lamblia (accession L29032.1) and Spironucleus salmonicida (accession KI546119.1) as an outgroup (not shown). Bootstrap values (1000 replicates) greater than 75% are shown on branches in red. Units for tree scale are inferred substitutions per base pair. Macaque-derived sequences (highlighted in orange) are named sequentially according to the animal from which they originated, e.g. Macaque1:1, Macaque1:2. The major Tetratrichomonas-like lineage of macaque-derived sequences is highlighted in red. Coloured dots indicate animal host taxa. Where available, Genbank accessions (Benson et al., 2015) are shown at the ends of tip labels.