Abstract
The correlation between ubiquitin-editing enzyme A20 and E3 ubiquitin ligase ring finger protein (RNF) 168 has been reported to be critical for repair of DNA damage. This study aimed to evaluate the potential role of this regulatory interaction in the pathogenesis of lupus nephritis (LN). The expression of RNF168 and A20 was measured in the podocytes derived from MRL/lpr murine lupus as well as patients with LN. Cell-based studies using renal podocytes bearing silenced RNF168, over-expressed A20, autophagy-related gene (Atg) 5 (a ubiquitin-like modifier), or silenced Atg5 were used to assess the effect of RNF168, A20, and Atg5 on DNA damage repair and nuclear factor kappa-B (NF-κB) activation in LN. It was found that podocyte autophagy was over-activated in LN and the abnormal podocyte autophagy led to down-regulation of A20, up-regulation of RNF168, and activation of the NF-κB. RNF168 silencing or A20 restoration inhibited activation of NF-κB pathway and promoted repair of DNA damage, where the level of autophagy was not changed. Activated A20 in podocytes weakened the promoting action of cell autophagy on RNF168. The current results suggest that RNF168 dysfunction may be involved in the pathogenesis of LN via down-regulation of A20 expression. Autophagy and RNF168 may be therapeutic targets for the prevention and treatment of LN.
Keywords: Lupus nephritis, A20, RNF168, NF-κB, Autophagy, Podocytes, DNA damage
Introduction
Systemic lupus erythematosus (SLE), characterized by unnecessary release of autoantibodies, especially antinuclear antibodies and antibodies against double-stranded DNA, is a relapsing autoimmune disorder that exerts great impact on several organs and tissues [1]. As one of the most prevalent and severe complications in patients with SLE, lupus nephritis (LN) presents renal patterns of damage, involving endothelial, tubulointerstitial, vascular, and most importantly, podocytes [2]. Possible mechanisms for podocyte injury consist of genetic factors, toxic injury, metabolic disturbance, as well as inflammatory response [3]. Recently, high expression of interferon alpha in patients has been reported to activate autophagy in both murine and human podocytes, while autophagy induction negatively correlates to podocyte injury [4].
The E3 ubiquitin ligase ring finger protein (RNF) 168 is a DNA damage response (DDR) factor that promotes histone 2A (H2A) ubiquitination and repair of DNA damage and accumulates in autophagy-defective cells [5]. More specifically, the ubiquitin-editing enzyme A20 (also termed as TNFAIP3) has been reported to modulate RNF168 expression, thus regulating tumor cell resistance to DNA damage therapy [6]. The close correlations between polymorphisms and mutations of A20 and inflammatory, autoimmune diseases and carcinoma have been validated [7]. Moreover, restoration of A20 possesses anti-inflammatory, antioxidant, anti-apoptotic, as well as pro-regenerative functions in hepatocytes, thereby serving as an extensive hepatoprotective goal [8]. A20 exerts its inhibitory functions by suppressing critical proinflammatory factors, such as nuclear factor kappa-B (NF-κB) [9]. Furthermore, it has also been confirmed that autophagy of macrophages contributes to the pathogenesis of SLE, enhancing the production of proinflammatory cytokines [10]. Based on the aforementioned findings, it was proposed that RNF168, A20, and autophagy may be involved in repair of DNA damage by regulating the expression of NF-κB in podocytes. Therefore, the aim of the present study was to assess whether the expression of RNF168 and A20 is altered in patients with LN and tissues of LN mice. Furthermore, the study investigated the correlation between RNF168 and A20 in podocytes and the mechanism of RNF168 and A20 protecting DNA against damage in podocytes. The observations of the present study may provide a theoretical underpinning in the prevention and therapy of LN.
Materials and Methods
Study Subjects
We collected renal biopsy specimens from 40 patients with LN (6 males and 34 females, aged 22–58 years) at the Xuzhou Central Hospital from January 2017 to September 2020 in this study. Twenty-five renal biopsy tissue samples from patients with nonrenal hematuria or paracancerous tissue samples from patients with renal carcinoma (6 males and 19 females at the age of 21–56 years) were collected as controls. There was no significant difference regarding the age and gender between the control samples and LN samples. All patients with LN fulfilled the American College of Rheumatology (ACR) 2009 classification criteria [11]. The urinalysis and renal biopsy confirmed renal tissue involvement. Patients who had taken immunosuppressive agents 2 months before the experiments were excluded. After isolation, the specimens were fixed with 10% paraformaldehyde at 4°C for 2–4 h, transferred into 30% sucrose phosphate-buffered saline (PBS) and reacted at 4°C. The study protocols were approved by the Biomedical Research Ethics Committee of Xuzhou Central Hospital (Approval Number: XZXY-LJ-20190222-005), and all participants provided written informed consents according to the Declaration of Helsinki.
Immunohistochemical Staining
The primary rabbit antibodies to RNF168 (ab235892, 1:800), and A20 (ab92324, 1:80), along with the horseradish peroxidase (HRP)-labeled secondary antibody to immunoglobulin (IgG) (ab6728, 1:100), were purchased from Abcam Inc. (Cambridge, UK). Normal saline was used as the negative control (NC) in replacement with the primary antibody. HRP-DAB Kit (P0203, Shanghai Beyotime Biotechnology Co. Ltd., Shanghai, China) was used for immunohistochemical staining. DAB is a common substrate for HRP. Under the catalysis of HRP, DAB produces brown precipitates. The protein expression was calculated by counting the number of positively stained cells in the total cells which were calculated in 5 randomly selected lesion areas under a microscope [12, 13].
RNA Isolation and Quantification
Total RNA was extracted from tissues or cells using prechilled TRIzol (Invitrogen Inc., Carlsbad, CA, USA). Then the total extracted RNA was reversely transcribed into complementary DNA (cDNA) using the cDNA reverse transcription kit (K1622, Reanta Co., Ltd., Beijing, China). Reverse transcription quantitative polymerase chain reaction (RT-qPCR) was carried out using the SYBR® Premix Ex TaqTM II kit (Takara, Dalian, Liaoning, China) on an ABI 7500 qPCR instrument (ABI Company, Oyster Bay, NY, USA). The primer sequences are listed in online supplementary Table 1 (for all online suppl. material, see www.karger.com/doi/10.1159/000527624). The relative mRNA expression was calculated using glyceraldehyde-3-phosphate dehydrogenase as an internal control (2−ΔΔCt method) [14].
Western Blot Analysis
The total protein was extracted from tissues or cells using radioimmunoprecipitation assay lysis buffer (R0010, Solarbio, Beijing, China) containing phenylmethanesulfonyl fluoride. The protein was separated by polyacrylamide gel electrophoresis and then transferred onto the polyvinylidene fluoride membrane by the wet transfer method. The membrane was blocked with 5% bovine serum albumin for 1 h at room temperature and incubated with diluted primary rabbit antibodies to RNF168 (ab106389, 1:2000), phosphorylated (p)-p65 (ab86299, phospho S536, 1:5000), total (t)-p65 (ab131100, 1:800), Atg5 (ab108327, 1:5000), light chain 3 (LC3)-II/LC3-I (ab51520, 1:3000), Beclin-1 (ab207612, 1:2000), A20 (ab74037, 1:800), and histone family member X (H2AX) phosphorylated on Ser139 (γH2AX) (ab11175, 1:5000) at 4°C overnight in a shaker. All antibodies were purchased from Abcam. After 3 washes with Tris-buffered saline Tween-20, the membranes were then incubated with the HRP-conjugated secondary goat anti-rabbit IgG (1:5000; ProteinTech Group, Chicago, IL, USA) for 1 h at room temperature. The relative protein level was quantified, as normalized to glyceraldehyde-3-phosphate dehydrogenase (10494-1-AP, 1:5000; ProteinTech), by the Quantity One v4.6.2 software.
Experimental Animals
The specific pathogen free-grade female MRL/MpJ-Faslpr (MRL/lpr) mice spontaneously developed lupus-like disease and MRL/MpJ mice as littermate control (n = 12, aged 8–10 weeks and weighing 18–22 g) were provided by Model Animal Research Center of Nanjing University (SCXK [Su] 2005-02, Nanjing, China). All mice were housed individually in a temperature- (24–26°C) and humidity-controlled (60–80%) room under a 12/12 h light/dark cycle with ad libitum access to chow and drinking water sterilized by high pressure. After 2 weeks, the mice were euthanized by anesthesia after fasting for 6–8 h and then weighed. The serum was collected, and the bilateral kidneys were weighed. A portion of renal tissues were fixed in neutral formalin for morphological examination, and the remaining was preserved in liquid nitrogen for further experiments. All animal procedures were approved by the Animal Ethics Committee of Southeast University (Approval Number: 201508002) and performed according to the Guide for the Care and Use of Laboratory Animals published by the US National Institutes of Health.
HE Staining
After fixed and dehydrated, the renal tissues were embedded and cut into 2–4-μm-thick sections. After conventional dewaxing, the sections were hydrated with gradient ethanol and then stained with hematoxylin for 5 min and with 0.5% eosin for 1 min. The pathological changes of the renal tissues were observed under an optical microscope. Five visual fields were taken from each section, and the ratio of area of inflammatory cell infiltration and necrosis to the total area in each field was calculated.
Periodic Acid-Schiff Staining
The renal tissues were dewaxed and oxidized with periodate. Following Schiff staining, the sections were stained with hematoxylin. The kidney injury was observed under a microscope. Renal pathological changes were assessed in a semiquantitative manner: Grade 0 indicated normal glomeruli with no renal interstitial infiltration; grade 1 indicated mild renal interstitial infiltration; grade 2 indicated moderate renal interstitial infiltration and mild to moderate glomerular hyperplasia; grade 3 indicated severe renal interstitial infiltration and severe glomerular hyperplasia with segmental or glomerular sclerosis or crescent formation [15, 16].
Urine Protein Content Measurement
At the end of the experiment, urine samples were collected with metabolic cages 24 h before the mice were euthanized. Coomassie brilliant blue G-250 (100 mg) was dissolved in 50 mL 95% ethanol, added with 100 mL phosphoric acid (0.85 g/mL), and diluted to 1,000 mL with distilled water. Bovine serum albumin (P0007; Baoman Biotechnology Co., Ltd, Shanghai, China) was used as the standard protein to plot a standard curve. A total of 0.1 mL sample was added with 0.9 mL distilled water, mixed thoroughly with 5 mL Coomassie brilliant blue G-250 protein reagent, and allowed to stand for 2 min. The optical density value was recorded at 595 nm.
Enzyme-Linked Immunosorbent Assay
According to the instructions of the enzyme-linked immunosorbent assay (ELISA) kit (YQ, Imunbio, Beijing, China), the serum IgG anti-dsDNA antibody (DKO095, DiaMetra, Perugia, Italy) was determined. The optical density value was measured at 492 nm with a microplate reader (BS-1101, Nanjing DeTie Laboratory Equipment Co., Ltd., Nanjing, Jiangsu, China). The standard curve was drawn and the content of anti-dsDNA antibody was calculated to evaluate renal inflammation.
Cell culture medium was collected from each group where the production of tumor necrosis factor-α (TNF-α) and interleukin (IL)-1β was measured in strict accordance with the instructions of Mouse TNF-α ELISA Kit (PT512, Beyotime) and Mouse IL-1β ELISA Kit (PI301, Beyotime). Each experiment was repeated three times.
Isolation and Culture of Mouse Podocytes
Podocytes were isolated from MRL/MpJ mice and MRL/lpr mice as previously described [17]. In short, the mice were anesthetized and subjected to Dynabeads cardiac perfusion. Next, the kidneys were taken out, peeled off, and cut into small pieces (1 mm3). Each pair of kidneys were digested in 1 mL digestion solution for 30 min in a 37°C water bath and stirred gently. Using the plunger of a 5 mL syringe as a pestle, the digestion solution was pressed through a 100 μm cell strainer on a 50 mL Falcon tube to separate the kidney glomeruli containing Dynabeads, which were then placed in 1 mL of culture medium. The kidney glomeruli were added to a Matrigel-coated P100 tissue culture dish containing 6 mL of cell culture medium, and cultured at 37°C in a 5% CO2 humidified atmosphere, allowing the kidney glomeruli to attach to the bottom of the culture dish (Fig. S1A). Once attached, the first cell to grow from the kidney glomeruli after approximately 3 days of culture was the podocyte (Fig. S1B). After 7 days of culture, most of the podocytes grew out of the kidney glomeruli and could be isolated by filtration (Fig. S1C). Thereafter, 2 mL 0.25% trypsin/ethylenediamine tetra-acetic acid was supplemented to separate cells and glomeruli, and 8 mL medium was added to inactivate trypsin, followed by filtration through a 30 μm cell strainer on a 50 mL Falcon tube. The kidney glomeruli should be on the top of the filter, and the podocytes were passed through the filter and collected in the filtrate. The podocytes were collected by centrifugation and cultured (Fig. S1D). The isolated podocytes can be cryopreserved in liquid nitrogen in a medium containing 5% dimethyl sulfoxide.
Podocyte identification was as follows: Wilms' tumor 1 (1:50, ab89901, Abcam, Cambridge, UK) expressed by podocytes, mature podocyte markers nephrin (1:100, PA5-20330, Thermo Fisher Scientific Inc., Waltham, MA, USA) and nestin (1:20, 14-5843-82, Thermo Fisher Scientific) and actin fibers (stained with phalloidin; A12379, Thermo Fisher Scientific) were detected by immunofluorescence staining (Fig. S2A–D). By selecting the fluorescence images of different visual fields, the purity of the isolated and purified podocytes was roughly counted, which was about 98%. The purity was calculated using the formula: purity (%) = number of positive fluorescent cells/total number of cells.
An immortalized podocyte cell line was derived from podocytes from MRL/MpJ and MRL/lpr mice transfected with retroviruses containing plpcx-SVtsa58 vector [18]. In order to maintain growth, undifferentiated podocytes were cultured on a plate coated with rat tail type I collagenase (Sigma-Aldrich Chemical Company, St Louis, MO, USA) and exposed to Roswell Park Memorial Institute (RPMI) 1,640 medium (Life Technology, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (FBS), 100 U/mL penicillin, and 100 mg/mL streptomycin at 37°C with 5% CO2. In order to promote differentiation, podocytes were cultured in RPMI 1640 medium with 10% FBS, 100 U/mL penicillin, and 100 mg/mL streptomycin at 37°C with 5% CO2 for 14 days. Some of these cells were replicated by limiting dilution. Other nephrotic positive clone cells were used in the following experiments.
Cell Treatment
The podocytes in the logarithmic growth period were seeded into six-well plates 24 h before transfection when the cell confluence reached about 70%. A total of 20 μL of Lipofectamine 2000 (11668019, Thermo Fisher Scientific) was diluted in 500 μL of serum-free medium and incubated for 5 min at room temperature. The plasmids and liposome were mixed gently and incubated for 20 min at room temperature. The cells were washed 3 times with serum-free medium and incubated with 2 mL serum-free medium in each well. The abovementioned liposome mixture was cultured for 6 h. The medium was renewed with 20% serum-containing Dulbeccos's Modified Eagle's Medium, and cells were collected for subsequent experiments after culture for 24–48 h. The podocytes were transfected with RNF168-short hairpin RNA (shRNA), pcDNA3-A20, or Atg5-shRNA, with NC-shRNA, pcDNA3 or pcDNA3-Atg5 (podocytes from MRL/MpJ mice), as well as the corresponding controls. All plasmids were purchased from Shanghai Genechem Co., Ltd. (Shanghai, China).
Reagents and Pharmacological Inhibitor Treatment
Autophagy inhibitor 3-methyladenine (3-MA; IM0190) and activator rapamycin (R8140) were purchased from Solarbio. After seeded into a 24-well plate at a density of 1.0 × 105 cells/mL, the immortalized podocytes from MRL/MpJ mice were stimulated with rapamycin (500 nM), and those from LN mice were subjected to 3-MA (5 mM) with untreated podocytes as controls.
Transfection of Green Fluorescent Protein-LC3 Plasmids
According to the instructions of green fluorescent protein (GFP)-LC3 lentivirus transfection provided by Shanghai Genechem Co., Ltd. (Shanghai, China), the podocytes of MRL/MpJ mice and MRL/lpr mice adherent to the wells were infected with lentivirus GFP-LC3 (multiplicity of infection = 10). After 24 h, the green fluorescence of podocytes was observed to detect the infection efficiency. Subsequently, the podocytes were screened by 2 μg/mL puromycin, and the stable podocyte cell line was seeded in a 12-well plate coated with 1% polylysine. After 24 h, the cells were fixed with 4% paraformaldehyde and stained with 4′,6-diamidino-2-phenylindole (DAPI). The images were finally obtained under a fluorescence microscope.
Immunofluorescence Assay
After conventional detachment and transfection, the cells of each group were counted and seeded in the immunofluorescence chamber at a density of 2 × 105 cells/well. When reaching about 90% confluence, the cells in each well were fixed with 4% paraformaldehyde, treated with 0.3% Triton, and blocked with goat serum. The cells were then probed with the primary antibodies to RNF168 (sc-101125, 1:200), A20 (ab74037, 1:800), p-p65 (ab86299, 1:500), Atg5 (ab108327, 1:200), or γH2AX (ab11175, 1:2500) overnight at 4°C. All antibodies were purchased from Abcam except for RNF168 (Santa Cruz Biotechnology Inc., Santa Cruz, CA, USA). The secondary antibody, fluorescein isothiocyanate-conjugated goat anti-rabbit (1:50; DAKO, Santa Clara, CA, USA) was added to the cells and incubated at room temperature for 1 h under conditions void of light. After stained with DAPI, the cells were sealed with anti-fluorescence quenching agent. The NC samples were subjected to the same steps with the primary antibody replaced by PBS. RNF168-, A20-, p-p65-, and γH2AX-positive nuclear cells and Atg5-positive cells were photographed under the fluorescence microscope. RNF168, A20, and Atg5 were stained red and p-p65 was green.
Neutral Comet Assay for DNA Damage Detection
Cell suspension was centrifuged at 1,000 r/min for 3 min, and the cell density was adjusted into 1 × 106 cells/mL with PBS. The comet assay was carried out with the reference to Singh and Stephens et al. [19]. Comets appeared was calculated and measured by Comet Assay Project 1.0.1 software. The DNA damage was quantified as the percentage of total DNA in the comet tail length and Olive tail moment (tail moment = tail DNA% × tail length). The results were compared with those of blank controls. The repair capacity of DNA double-strand breaks was calculated according to the formula:

where M12 indicates mean value of Olive tail moment at the 12th h after quartz action, M24 indicates the mean Olive tail moment at the 24th h after quartz action, and M0 indicates the mean value of Olive tail moment without the quartz action.
Statistical Analysis
The data were analyzed by the SPSS 21.0 statistical software (IBM Corp, Armonk, NY, USA). All data are presented as mean ± standard deviation. Data with normal distribution and homogeneity of variance between two groups were compared using unpaired t test. Repeated measures analysis of variance with Tukey's post hoc test was applied for the comparison of data among multiple groups. p values under 0.05 were considered statistically significant.
Results
RNF168 Is Up-Regulated in Renal Tissues of LN Patients and Podocytes of LN Mice
We initially developed a mouse LN model and observed the appearance of MRL-lpr/lpr (MRL/lpr) (LN) and MRL/MpJ (control) mice, finding that the neck of LN mice was significantly larger than that of control mice (online suppl. Fig. S3A). In addition, there was no significant change in body weight and kidney weight between LN and control mice (online suppl. Table 2). Histopathological analysis on renal tissues by HE staining suggested tubular epithelial cell necrosis and tubulointerstitial fibrosis accompanied by infiltrated inflammatory cells in LN mice. Additionally, the pathological score of renal tissues of LN mice was much higher than that of control mice (online suppl. Fig. S3B). Periodic acid-Schiff staining results showed that the degree of renal tissue injury in LN mice was higher than that in control mice (online suppl. Fig. S3C). Furthermore, the 24-h urine protein content in the LN mice was elevated, relative to that in control mice (online suppl. Fig. S3D). The level of serum IgG-ds-DNA in LN mice was found to be increased versus that in control mice (online suppl. Fig. S3E). The above results indicated that MRL/lpr mice had severe kidney damage, which was in line with LN-related expectations. MRL/lpr mice were suitable for LN animal models.
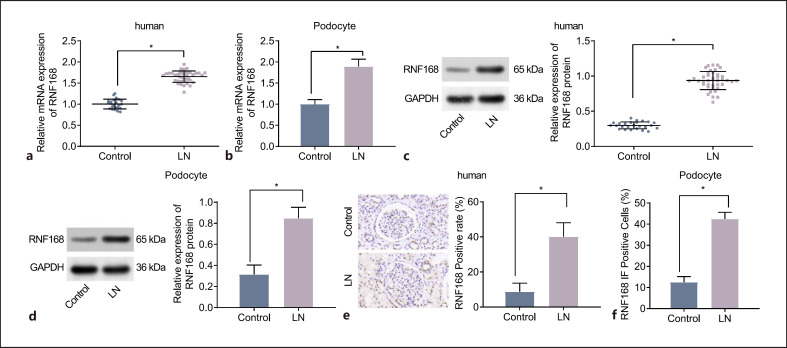
In addition, the RT-qPCR and Western blot results showed that the expression of RNF168 was higher in both renal tissues of LN patients (Fig. 1a, c) and podocytes of LN mice (Fig. 1b, d) than that in their separate controls. Immunohistochemistry detection revealed that RNF168 was mainly located in the nucleus, and its positive expression (brown-stained granules) was higher in human LN renal tissues than that in control renal tissues (Fig. 1e). These results suggested that RNF168 was located in the nucleus and highly expressed in LN tissues. Finally, the immunofluorescence analysis illustrated that the positive expression of RNF168 was mainly located in the nucleus. The expression of RNF168 was increased in the podocytes of the LN mice compared to that in the podocytes of control mice (Fig. 1f). These results suggested that RNF168 positive expression was located in the nucleus of podocytes and expressed at a high level in LN.
Fig. 1.
RNF168 is expressed at high levels in LN renal tissues and podocytes. a mRNA expression of RNF168 in control tissues (n = 25) and LN renal tissues (n = 40) determined by RT-qPCR. b mRNA expression of RNF168 in podocytes of control and LN mice measured by RT-qPCR. c Protein expression of RNF168 in control tissues (n = 25) and LN renal tissues (n = 40) determined by Western blot analysis. d Protein expression of RNF168 in podocytes of control and LN mice determined by Western blot analysis. e Immunohistochemistry of the positive expression of RNF168 protein in control tissues (n = 25) and LN renal tissues (n = 40). f Immunofluorescence staining of RNF168 in renal podocytes of control and LN mice. n = 12 for mice in each group. *p < 0.05 versus control tissues or control mice.
Inhibition of RNF168 Promotes Repair of DNA Damage of Podocytes in LN Mice
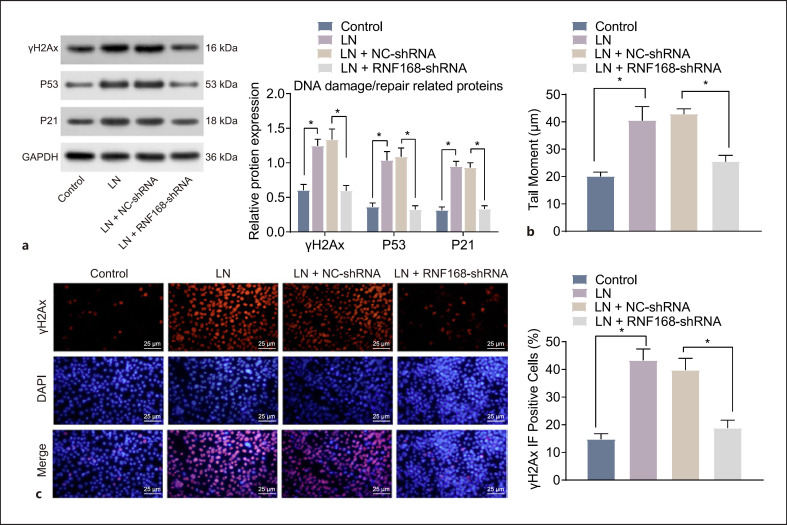
RNF168 was knocked down in the podocytes of MRL/lpr mice to investigate its effect on DNA repair of podocytes in LN. The results of RT-qPCR and Western blot analysis revealed that RNF168 was efficiently knocked down by the RNF168-shRNA plasmid, where the expression of RNF168 was markedly reduced in podocytes transfected with RNF168-shRNA (online suppl. Fig. S4A, B). Subsequently, the Western blot analysis revealed that in comparison with the podocytes from MRL/MpJ mice, the expression of DNA damage/repair-related proteins (γH2Ax, p53, and p21) was increased significantly in the podocytes from MRL/lpr. However, when RNF168 was knocked down, the levels of these proteins were notably decreased (Fig. 2a). Furthermore, the podocytes of LN mice exhibited significantly increased tail length, Olive tail moment, and rate of tail DNA compared to control mice. The podocytes treated with NC-shRNA showed consistent results to the podocytes of LN mice. However, the podocytes of LN mice, in response to RNF168 silencing, presented with significantly decreased tail length, Olive tail moment, and rate of tail DNA (Fig. 2b), suggesting that DNA was repaired. Finally, immunofluorescence results showed significantly higher γH2Ax positive nuclear expression in LN mouse podocytes than control mouse podocytes. Compared with NC-shRNA-treated podocytes of LN mice, γH2Ax positive nuclear expression showed a significant decline after RNF168-shRNA treatment (Fig. 2c). These data demonstrated that inhibition of RNF168 could facilitate the repair of DNA damage in the podocytes of LN mice.
Fig. 2.
Repair of DNA damage is promoted by RNF168 silencing in the podocytes of LN mice. The podocytes of MRL/lpr mice were transfected with RNF168-shRNA plasmids, with those transfected with NC-shRNA plasmids as the control. Podocytes of MRL/lpr mice without transfection were evaluated with those untreated podocytes from MRL/MpJ mice as the control. a Protein expression of γH2Ax, p53, and p21 in the podocytes at 24 h after treatment determined by Western blot analysis. b DNA damage pattern and tail moment in the podocytes at 24 h after treatment detected by the neutral comet assay. c Immunofluorescence staining of γH2Ax positive nuclear expression in the podocytes at 24 h after treatment. Cell experiment was repeated 3 times independently. *p < 0.05 versus the podocytes from MRL/MpJ mice or the podocytes transfected with NC-shRNA plasmids.
Inhibition of RNF168 Represses the NF-κB Signaling Pathway Activation
In order to study the activation of NF-κB signaling pathway in podocytes of LN mice, we evaluated the expression of NF-κB signaling pathway activation (p-p65 and t-p65) and downstream target genes, TNF-α and IL-1β. The results showed that compared with the podocytes from MRL/MpJ mice, the protein expression of p-p65 was significantly elevated in the untreated podocytes from MRL/lpr mice. In comparison with the podocytes of LN mice treated with NC-shRNA, the protein expression of p-p65 was decreased significantly following RNF168-shRNA treatment, but there was no significant difference in the t-p65 protein expression (Fig. 3a). Next, the ELISA results presented significantly higher levels of TNF-α and IL-1β production in the culture medium of untreated podocytes from MRL/lpr mice than those in the podocytes from MRL/MpJ mice, while they were reduced in RNF168-shRNA-treated podocytes versus that in NC-shRNA-treated podocytes (Fig. 3b). The immunofluorescence staining further suggested that p-p65 was mainly located in the cytoplasm and RNF168 was in the nuclei of podocytes from MRL/MpJ mice. In untreated podocytes from MRL/lpr mice, the numbers of RNF168- and p-p65-positive cells translocated into nuclei were elevated. When RNF168 was downregulated, the expression of RNF168 as well as the number of cells with p-p65 into nuclei was reduced (Fig. 3c). Together, these observations indicated that inhibition of RNF168 expression could hinder activation of the NF-κB signaling pathway.
Fig. 3.
The activation of the NF-κB signaling pathway is disrupted by the inhibition of RNF168 expression in podocytes of LN mice. a Protein expression of p-p65 (phospho S536) and t-p65 in the podocytes assessed by Western blot analysis. b Levels of TNF-α and IL-1β in the culture medium of podocytes assessed by ELISA. c Immunofluorescence staining of RNF168 and p-p65 (phospho S536) translocation into the nuclei of the podocytes. Cell experiment was repeated 3 times independently. *p < 0.05 versus the podocytes from MRL/MpJ mice or podocytes transfected with NC-shRNA plasmids.
Inhibition of Autophagy of Podocytes Restores the Expression of A20 and Represses the RNF168 Translocation into Nuclei, Thus Promoting the Repair of DNA Damage
In order to explore the effect of autophagy on podocytes of LN mice, the podocytes of MRL/MpJ mice or MRL/lpr mice were used as research subjects. Initially, the renal podocytes of MRL/MpJ mice were treated with rapamycin reagent to activate autophagy, while renal podocytes from MRL/lpr mice were added with 3-MA to inhibit cell autophagy. Then, pcDNA3-Atg5 plasmids were transfected into the podocytes of MRL/MpJ mice to activate autophagy, and Atg5-shRNA plasmids were transfected into the podocytes of MRL/lpr mice to inhibit autophagy.
According to Western blot analysis, compared with untreated podocytes from MRL/MpJ mice, rapamycin (online suppl. Fig. S5A) and pcDNA3-Atg5 (online suppl. Fig. S5C) promoted the protein expression of autophagy-related proteins (Atg5, LC3-II/LC3-I, and Beclin-1). Meanwhile, treatment with 3-MA (online suppl. Fig. S5A) and Atg5-shRNA (online suppl. Fig. S5C) inhibited the protein expression of Atg5, LC3-II/LC3-I, and Beclin-1.
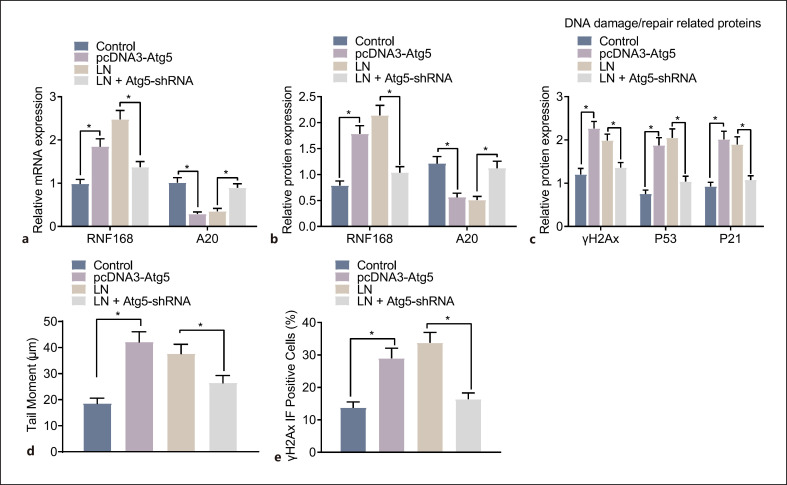
Furthermore, autophagy was also detected by GFP-LC3 labeling and Atg5 immunofluorescence, where GFP-LC3 showed green fluorescence, Atg5 showed red fluorescence, and the nuclei were stained blue by DAPI. Rapamycin (online suppl. Fig. S5B) and pcDNA3-Atg5 (online suppl. Fig. S5D) in the podocytes from MRL/MpJ mice led to promoted autophagy as evidenced by increased GFP-LC3/Atg5 cells, whereas 3-MA (online suppl. Fig. S5B) and Atg5-shRNA (online suppl. Fig. S5D) in the podocytes from MRL/lpr mice contributed to suppressed autophagy as presented with decreased GFP-LC3/Atg5 cells. Moreover, the RT-qPCR and Western blot analysis indicated that rapamycin (online suppl. Fig. S6A, B) and pcDNA3-Atg5 (Fig. 4a, b) in the podocytes from MRL/MpJ mice induced A20 degradation and promoted the accumulation of RNF168. As expected, 3-MA (online suppl. Fig. S6A, B) and Atg5-shRNA (Fig. 4a, b) in the podocytes from MRL/lpr mice resulted in the opposite trends.
Fig. 4.
Podocyte autophagy inhibition restores the expression of A20 and blocks the RNF168 translocation into nuclei, contributing to the repair of DNA damage. Podocytes from MRL/MpJ mice were treated with pcDNA3-Atg5 plasmids, and podocytes from MRL/lpr mice were treated with Atg5-shRNA plasmids. a mRNA expression of RNF168 and A20 in the podocytes evaluated by RT-qPCR. b Protein expression of RNF168 and A20 in the podocytes evaluated by Western blot analysis. c Protein expression of γH2Ax, p53, and p21 in the podocytes evaluated by Western blot analysis. d DNA damage pattern and tail moment in the podocytes at 24 h after treatment detected by the neutral comet assay. e Immunofluorescence staining of γH2Ax positive nuclear expression in the podocytes at 24 h after treatment. Cell experiment was repeated 3 times independently. *p < 0.05 versus untreated podocytes from MRL/MpJ mice or MRL/lpr mice.
Moreover, enhanced protein expression of DNA damage/repair-associated proteins (γH2Ax, p53, and p21) was observed after treatment of rapamycin (online suppl. Fig. S6C) or pcDNA3-Atg5 (Fig. 4c). By contrast, the expression of γH2Ax, p53, and p21 was decreased following treatment of 3-MA (online suppl. Fig. S6C) or Atg5-shRNA (Fig. 4c). The following neutral comet assay and immunofluorescence detection suggested that treatment of rapamycin (online suppl. Fig. S6D, E) or pcDNA3-Atg5 (Fig. 4d, e) was responsible for the enhanced tail moment and γH2Ax-positive nuclear cells. Thus, inhibition of autophagy of podocytes could restore the expression of A20 and inhibit the RNF168 translocation into nuclei, thereby inducing the repair of DNA damage.
Inhibited Podocyte Autophagy Downregulates RNF168 Expression and Consequently Blocks the NF-κB Signaling Pathway Activation
We next moved to study the effect of autophagy on the activity of NF-κB in podocytes of LN mice. The Western blot and ELISA results presented that, in response to treatment with rapamycin (Fig. 5a, b) or the pcDNA3-Atg5 plasmids (Fig. 5d, e), the protein expression of p-p65 and levels of TNF-α and IL-1β were elevated, relative to untreated podocytes from MRL/MpJ mice, while they were diminished in response to treatment with 3-MA (Fig. 5a, b) or Atg5-shRNA plasmids (Fig. 5d, e) versus the untreated podocytes from MRL/lpr mice. Besides, no significant difference was noted in the protein expression of t-p65 (p > 0 05) (Fig. 5a). Finally, immunofluorescence detection further observed that p-p65 was located in the cytoplasm and RNF168 in the nuclei in untreated podocytes from MRL/MpJ mice (Fig. 5c, f). The RNF168- and p-p65-positive cells after the treatment of rapamycin or pcDNA3-Atg5 were significantly elevated. Consistently, RNF168- and p-p65-positive cells were reduced after treatment with 3-MA or Atg5-shRNA plasmids. These findings suggested that inhibition of podocyte autophagy could down-regulate the expression of RNF168 and thus block the activation of the NF-κB signaling pathway.
Fig. 5.
The expression of RNF168 and the activation of NF-κB signaling pathway are impaired by inhibition of podocyte autophagy. Protein expression of p-p65 (phospho S536) and t-p65 in podocytes assessed by Western blot analysis in response to 3-MA (a) or Atg5-shRNA (d). Levels of TNF-α and IL-1β in the culture medium of podocytes assessed by ELISA in response to 3-MA (b) or Atg5-shRNA (e). Immunofluorescence staining of RNF168 and p-p65 (phospho S536) in podocytes in response to 3-MA (c) or Atg5-shRNA (f). Cell experiment was repeated 3 times independently. *p < 0.05 versus untreated podocytes from MRL/MpJ mice or MRL/lpr mice.
Activation of A20 Represses the RNF168 Translocation into Nuclei in the Podocytes of LN Mice, Thus Promoting the Repair of DNA Damage
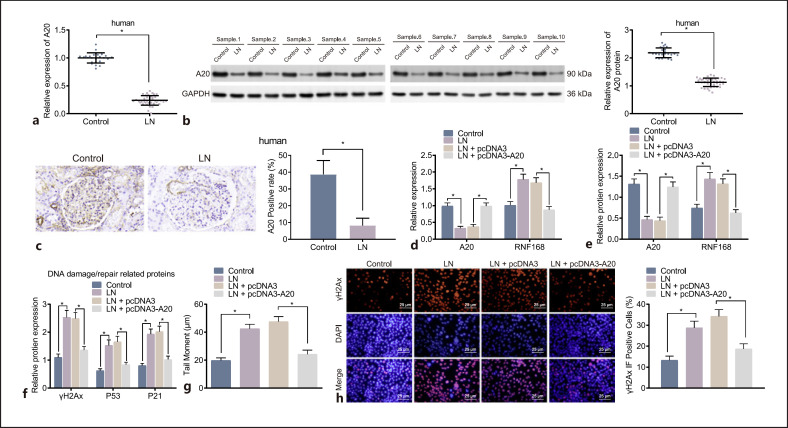
Further investigation was performed to explore the effect of A20-mediated RNF168 on DNA damage repair in podocytes of LN mice. The RT-qPCR and Western blot analysis displayed that the expression of A20 was lower in renal tissues of LN patients than that in control renal tissues (Fig. 6a, b). Additionally, immunohistochemistry results showed that A20 protein-stained yellow brown was expressed in the cytoplasm, nucleus, and cell membrane of podocytes. Meanwhile, the positive expression of A20 protein was downregulated in LN patient renal tissues relative to that in control renal tissues (Fig. 6c). These results indicated that A20 was expressed at a low level in renal tissues of LN patients and expressed in the cytoplasm, nucleus, and cell membrane.
Fig. 6.
The accumulation of RNF168 is abrogated and the repair of DNA damage is accelerated by up-regulation of A20. a mRNA expression of A20 in control tissues (n = 25) and LN renal tissues (n = 40) determined by RT-qPCR. b Protein expression of A20 in control tissues (n = 25) and LN renal tissues (n = 40) determined by Western blot analysis. c Immunohistochemical staining of positive expression of A20 protein in control tissues (n = 25) and LN renal tissues (n = 40). d mRNA expression of RNF168 and A20 in podocytes in response to pcDNA3 or pcDNA3-A20 evaluated by RT-qPCR. e Protein expression of RNF168 and A20 in podocytes evaluated by Western blot analysis. f Protein expression of γH2Ax, p53, and p21 in podocytes in response to pcDNA3 or pcDNA3-A20 evaluated by Western blot analysis. g DNA damage pattern and tail moment in the podocytes in response to pcDNA3 or pcDNA3-A20 at 24 h after treatment measured by the neutral comet assay. h Immunofluorescence staining of γH2Ax positive nuclear expression in the podocytes in response to pcDNA3 or pcDNA3-A20 at 24 h after treatment. Cell experiment was repeated 3 times independently. *p < 0.05 versus untreated podocytes from MRL/MpJ mice or MRL/lpr mice.
The effects of A20 on the RNF168 expression and repair of DNA damage were explored. The podocytes of MRL/lpr mice were transfected with pcDNA3-A20, and the expression of RNF168 and A20 in the podocytes was measured using RT-qPCR and Western blot analysis. Compared with podocytes of MRL/MpJ mice, the expression of RNF168 was increased significantly, while that of A20 was decreased significantly in MRL/lpr mice. In addition, over-expression of A20 inhibited RNF168 expression (Fig. 6d, e). Further Western blot analysis suggested that protein expression of γH2Ax, p53, and p21 was downregulated in cells after over-expression of A20 (Fig. 6f). The following results of comet assay and immunofluorescence suggested that over-expression of A20 was responsible for reduced tail moment and γH2Ax-positive nuclear cells (Fig. 6g, h). Overall, these results suggested that up-regulation of A20 could repress accumulation of RNF168 and potentiate the repair of DNA damage in podocytes of LN mice.
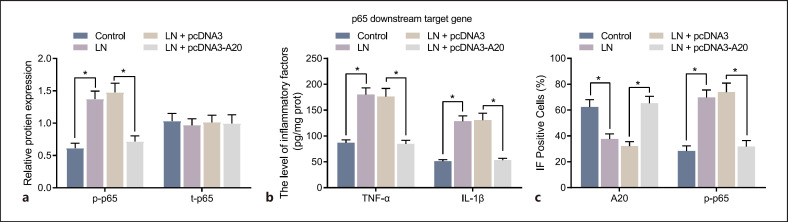
Restoration of A20 Represses Activation of the NF-κB Signaling Pathway in the Podocytes of LN Mice
In order to study the effect of A20 on the activation of the NF-κB signaling pathway, the expression of p-p65 (phospho S536), and t-p65, along with the levels of p65 downstream target genes (TNF-α and IL-1β) in the culture medium of podocytes was detected using Western blot analysis and ELISA, respectively. It was found that the expression of t-p65 was not statistically significant in the presence of A20 up-regulation, while (phospho S536) p-p65 expression, and TNF-α, and IL-1β levels induced in MRL/lpr mice were suppressed following over-expression of A20 (Fig. 7a, b). The results of immunofluorescence staining showed that the expression of p-p65 (phospho S536) was mainly located in the cytoplasm and A20 in both the nucleus and cytoplasm of podocytes from MRL/MpJ mice. Nevertheless, the number of nuclear A20-positive cells in untreated podocytes MRL/lpr mice was significantly lower than that in MRL/MpJ mice, while the number of nuclear p-p65 (phospho S536)-positive cells was significantly increased (Fig. 7c). In MRL/lpr mice, the number of nuclear A20-positive cells was significantly enhanced in the podocytes treated with pcDNA3-A20, while that of nuclear p-p65 (phospho S536)-positive cells was significantly reduced (Fig. 7c). These results suggested that activated A20 could inactivate the NF-κB signaling pathway in the podocytes of LN mice.
Fig. 7.
Over-expression of A20 inactivates the NF-κB signaling pathway in the podocytes of LN mice. a Protein expression of p-p65 (phospho S536) and t-p65 in the podocytes assessed by Western blot analysis. b Levels of TNF-α and IL-1β in the culture medium of podocytes assessed by ELISA. c Immunofluorescence staining of A20 and p-p65 (phospho S536) in the podocytes. Cell experiment was repeated 3 times independently. *p < 0.05 versus untreated podocytes from MRL/MpJ mice or MRL/lpr mice.
Discussion
Autophagy plays an essential role in the immune system and is involved in autoimmune diseases; there is compelling evidence suggesting the relationship between autophagy and LN, known as a systemic autoimmune disorder [20]. In the present study, we clarified that the augmented autophagy inhibited A20 expression, thus up-regulating RNF168 expression, promoting DNA damage, and activating the NF-κB signaling pathway, ultimately exacerbating LN.
The localization of RNF168 at the site of damage has been shown to highly increase the local concentration of ubiquitinated proteins and determine the prolonged ubiquitination signal [21]. Specifically, RNF168 is localized in the nucleus to play a key role of ubiquitination in DDR, and in several DDR processes, RNF168 localizes at repair of DNA damage sites and promotes mono-ubiquitination of H2A/H2AX at K13-15 to induce repair of DNA damage complex formation [22]. Our study first revealed that RNF168 was mainly located in the nucleus of podocytes and its expression was found to be higher in tissue samples from patients with LN and podocytes of LN mice than that in the healthy controls. In addition, inhibition of RNF168 could promote repair of DNA damage. Muñoz et al. [23] confirmed that the change of RNF168(R57) is necessary for several DDR functions, including impairment of homologous recombination, induction of the ubiquitination of K13/15 on H2Ax. Ubiquitin-specific protease 14, a deubiquitinase, has been reported to facilitate the repair of DNA damage by targeting RNF168-dependent ubiquitination [24]. Another study conducted by Shire et al. [25] validated that silencing of RNF168 can elevate nuclear bodies of promyelocytic leukemia, which results in apoptosis, p53 activation, as well as the repair of DNA damage. Ubiquitination confers significant effect on NF-κB activation, and ubiquitylated transmitters of NF-κB signaling can accumulate in proximity to endomembranes, such as RNF121 [26] and RNF183 [27], both of which have been revealed as novel players implicated in the signaling contributing to NF-κB activation. In this study, inhibition of RNF168 was observed to block activation of the NF-κB signaling pathway.
Moreover, inhibition of autophagy could decrease the expression of RNF168, and restore A20 expression, thereby blocking the activation of NF-κB signaling pathway and accelerating repair of DNA damage in podocytes from LN mice. Autophagy has been implicated in many repair of DNA damage mechanisms, such as base excision repair, nucleotide excision repair, as well as mismatch repair along with the extensively reported autophagy positive roles on homologous recombination [28]. Different from membrane receptor-initiated signaling pathways, the vital signaling initiation event stimulated by genotoxic agents contributing to NF-κB signaling pathway activation remain completely understudied [29]. Autophagy has been found to sequester A20 to promote the NF-κB signaling pathway activation in macrophages [30]. Shi et al. [31] indicated that A20 exerts an inhibitory role over lipopolysaccharide-stimulated autophagy in macrophages by reducing the extent of ubiquitination of Beclin-1, suggesting the adverse relationship between A20 and autophagy. Moreover, SQSTM1, an autophagy substrate and receptor for degradation of ubiquitinated substrates via autophagy, binds to RNF168 and reduces its E3 ligase activity [32]. Furthermore, RNF168-associated ubiquitination signaling is reduced in autophagy-deficient cells [24]. Another report demonstrates that depletion of SQSTM1 in lymphoblastic cell lines results in remarkable promotion of endogenous RNF168-γH2Ax damage foci in addition to chromatin ubiquitination, suggesting the induction of RNF168-controlled repair of DNA damage mechanism [33].
We also found that induction of A20 decreased RNF168 accumulation and disrupted the NF-κB signaling pathway activation, thus promoting the repair of DNA damage. In peripheral blood mononuclear cells, the down-regulation of A20 gene has been observed, which involves in the pathogenesis of the SLE disease [34]. Loss of A20 in dendritic cells may contribute to spontaneously occurring colitis, seronegative arthritis, and enthesitis in mice, which implies that disturbance of A20-dependent dendritic cells underlies colitis and colitis-related arthritis in clinical samples [35]. The ubiquitin-editing enzyme A20/TNFAIP3 has been reported to bind and inhibit the E3 ubiquitin ligase RNF168, which is responsible for the regulation of histone H2A turnover critical for proper repair of DNA damage [6]. A20, a key negative regulator of inflammation [36], has long been validated to participate in the termination of the NF-κB signaling pathway which is engaged in the pathogenesis of SLE [37]. More specifically, A20 over-expression can exert protective effects on podocyte injury in LN by impairing the activation of the NF-κB signaling pathway [38]. In partially accordance with our results, treatment with A20 over-expression can significantly alleviate pristine-induced lupus inflammation and renal injury by inhibiting the NF-κB and NLRP3 inflammasome activation in macrophages in mice [36].
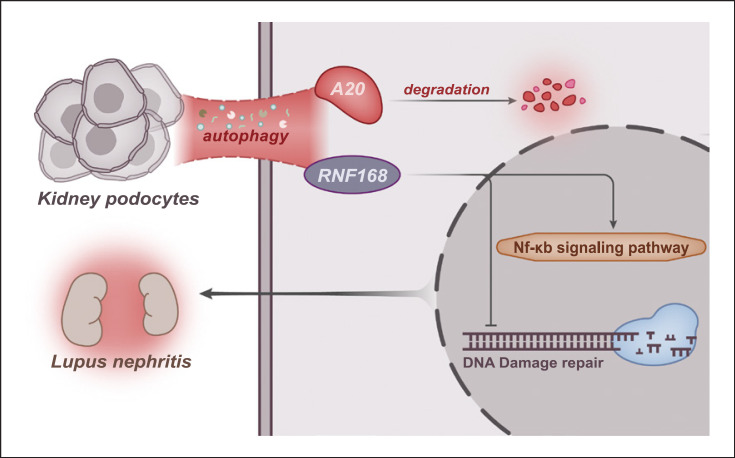
Overall, our findings illuminated that activated A20 due to the inhibited autophagy could potentially decrease RNF168 accumulation and inhibit the NF-κB signaling pathway, thus enhancing the repair of DNA damage and improving LN phenotypes (Fig. 8). This novel mode may represent a more efficient treatment option for future LN therapy. However, the current study only presents the theoretical basis of this mechanism and further studies should be performed in animal models, so as to support a promising clinical application in the treatment for patients with LN.
Fig. 8.
Schematic diagram of the mechanism by which podocyte autophagy affects the repair of DNA damage and LN progression. Podocyte autophagy promotes the degradation of A20 and the RNF168 translocation into nuclei, activating the NF-κB signaling pathway, suppressing the repair of DNA damage, and aggravating LN.
Statement of Ethics
The study protocols were approved by the Biomedical Research Ethics Committee of Xuzhou Central Hospital (Approval Number: XZXY-LJ-20190222-005), and all participants provided written informed consents according to the Declaration of Helsinki. All animal procedures were approved by the Animal Ethics Committee of Southeast University (Approval Number: 201508002) and performed according to the Guide for the Care and Use of Laboratory Animals published by the US National Institutes of Health.
Conflict of Interest Statement
The authors declare that they have no conflict of interest.
Funding Sources
This study was supported by National Natural Science Foundation of China (81600540), Key project of Jiangsu Commission of Health (ZD2022044), Natural Science Foundation of Jiangsu Province (BK20150224), Science and Technology Foundation of Xuzhou City (KC20182, KC21186), Science and Technology Foundation of Xuzhou Health Committee (XWKYHT20200020), Jiangsu Six Talent Peaks Project, Jiangsu Entrepreneurial Innovation Program, Xuzhou Entrepreneurial Innovation Program, and Open Project of Key Laboratories in Jiangsu Province (XZSYSKF2021031).
Author Contributions
Luxi Zou, Ling Sun, Ruixue Hua, Yu Wu, Linlin Sun, and Ting Chen designed the study. Luxi Zou and Ling Sun contributed to drafting the manuscript. Ruixue Hua and Yu Wu collated the data and designed and developed the database. Linlin Sun and Ting Chen contributed to check and edit the whole manuscript. All authors approved the final manuscript.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Supplementary Material
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Funding Statement
This study was supported by National Natural Science Foundation of China (81600540), Key project of Jiangsu Commission of Health (ZD2022044), Natural Science Foundation of Jiangsu Province (BK20150224), Science and Technology Foundation of Xuzhou City (KC20182, KC21186), Science and Technology Foundation of Xuzhou Health Committee (XWKYHT20200020), Jiangsu Six Talent Peaks Project, Jiangsu Entrepreneurial Innovation Program, Xuzhou Entrepreneurial Innovation Program, and Open Project of Key Laboratories in Jiangsu Province (XZSYSKF2021031).
References
- 1.Gregersen JW, Jayne DRW. B-cell depletion in the treatment of lupus nephritis. Nat Rev Nephrol. 2012;8((9)):505–514. doi: 10.1038/nrneph.2012.141. [DOI] [PubMed] [Google Scholar]
- 2.Wang Y, Yu F, Song D, Wang SX, Zhao MH. Podocyte involvement in lupus nephritis based on the 2003 ISN/RPS system a large cohort study from a single centre. Rheumatology. 2014;53((7)):1235–1244. doi: 10.1093/rheumatology/ket491. [DOI] [PubMed] [Google Scholar]
- 3.Cellesi F, Li M, Rastaldi MP. Podocyte injury and repair mechanisms. Curr Opin Nephrol Hypertens. 2015;24((3)):239–244. doi: 10.1097/MNH.0000000000000124. [DOI] [PubMed] [Google Scholar]
- 4.Zhou XJ, Klionsky DJ, Zhang H. Podocytes and autophagy a potential therapeutic target in lupus nephritis. Autophagy. 2019;15((5)):908–912. doi: 10.1080/15548627.2019.1580512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang Y, Zhang N, Zhang L, Li R, Fu W, Ma K, et al. Autophagy regulates chromatin ubiquitination in DNA damage response through elimination of SQSTM1/p62. Mol Cell. 2016;63((1)):34–48. doi: 10.1016/j.molcel.2016.05.027. [DOI] [PubMed] [Google Scholar]
- 6.Yang C, Zang W, Tang Z, Ji Y, Xu R, Yang Y, et al. A20/TNFAIP3 regulates the DNA damage response and mediates tumor cell resistance to DNA-damaging therapy. Cancer Res. 2018;78((4)):1069–1082. doi: 10.1158/0008-5472.CAN-17-2143. [DOI] [PubMed] [Google Scholar]
- 7.Ma A, Malynn BA. A20 linking a complex regulator of ubiquitylation to immunity and human disease. Nat Rev Immunol. 2012;12((11)):774–785. doi: 10.1038/nri3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.da Silva CG, Cervantes JR, Studer P, Ferran C. A20 an omnipotent protein in the liver: prometheus myth resolved? Adv Exp Med Biol. 2014;809:117–139. doi: 10.1007/978-1-4939-0398-6_8. [DOI] [PubMed] [Google Scholar]
- 9.Verstrepen L, Verhelst K, van Loo G, Carpentier I, Ley SC, Beyaert R. Expression biological activities and mechanisms of action of A20 (TNFAIP3) Biochem Pharmacol. 2010;80((12)):2009–2020. doi: 10.1016/j.bcp.2010.06.044. [DOI] [PubMed] [Google Scholar]
- 10.Li B, Yue Y, Dong C, Shi Y, Xiong S. Blockade of macrophage autophagy ameliorates activated lymphocytes-derived DNA induced murine lupus possibly via inhibition of proinflammatory cytokine production. Clin Exp Rheumatol. 2014;32((5)):705–714. [PubMed] [Google Scholar]
- 11.Schwartz N, Goilav B, Putterman C. The pathogenesis diagnosis and treatment of lupus nephritis. Curr Opin Rheumatol. 2014;26((5)):502–509. doi: 10.1097/BOR.0000000000000089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Trere D, Montanaro L, Ceccarelli C, Barbieri S, Cavrini G, Santini D, et al. Prognostic relevance of a novel semiquantitative classification of Bcl2 immunohistochemical expression in human infiltrating ductal carcinomas of the breast. Ann Oncol. 2007;18((6)):1004–1014. doi: 10.1093/annonc/mdm074. [DOI] [PubMed] [Google Scholar]
- 13.Ellis IO, Bartlett J, Dowsett M, Humphreys S, Jasani B, Miller K. Best practice No 176 updated recommendations for HER2 testing in the UK. J Clin Pathol. 2004;57((3)):233–237. doi: 10.1136/jcp.2003.007724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ayuk SM, Abrahamse H, Houreld NN. The role of photobiomodulation on gene expression of cell adhesion molecules in diabetic wounded fibroblasts in vitro. J Photochem Photobiol B. 2016;161:368–374. doi: 10.1016/j.jphotobiol.2016.05.027. [DOI] [PubMed] [Google Scholar]
- 15.Xu YX, Tan Y, Yu F, Zhao MH. Late onset lupus nephritis in Chinese patients classified by the 2003 international society of nephrology and renal pathology society system. Lupus. 2011;20((8)):801–808. doi: 10.1177/0961203310397563. [DOI] [PubMed] [Google Scholar]
- 16.Silva GEB, Costa RS, Ravinal RC, Ramalho LZ, Dos Reis MA, Coimbra TM. NF-kB expression in IgA nephropathy outcome. Dis Markers. 2011;31((1)):9–15. doi: 10.3233/DMA-2011-0795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vasilopoulou E. Isolating and culturing mouse podocyte cells to study diabetic nephropathy. Methods Mol Biol. 2020;2067:53–59. doi: 10.1007/978-1-4939-9841-8_5. [DOI] [PubMed] [Google Scholar]
- 18.Jat PS, Sharp PA. Cell lines established by a temperature-sensitive simian virus 40 large-T-antigen gene are growth restricted at the nonpermissive temperature. Mol Cell Biol. 1989;9((4)):1672–1681. doi: 10.1128/mcb.9.4.1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Souliotis VL, Sfikakis PP. Increased DNA double-strand breaks and enhanced apoptosis in patients with lupus nephritis. Lupus. 2015;24((8)):804–815. doi: 10.1177/0961203314565413. [DOI] [PubMed] [Google Scholar]
- 20.Wang L, Law HK. The role of autophagy in lupus nephritis. Int J Mol Sci. 2015;16((10)):25154–67. doi: 10.3390/ijms161025154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pinato S, Scandiuzzi C, Arnaudo N, Citterio E, Gaudino G, Penengo L. RNF168 a new RING finger MIU-containing protein that modifies chromatin by ubiquitination of histones H2A and H2AX. BMC Mol Biol. 2009;10((1)):55. doi: 10.1186/1471-2199-10-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu N, Xue M, Wang W, Xia D, Li Y, Zhou X, et al. RNF168 facilitates proliferation and invasion of esophageal carcinoma possibly via stabilizing STAT1. J Cell Mol Med. 2019;23((2)):1553–1561. doi: 10.1111/jcmm.14063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Munoz MC, Yanez DA, Stark JM. An RNF168 fragment defective for focal accumulation at DNA damage is proficient for inhibition of homologous recombination in BRCA1 deficient cells. Nucleic Acids Res. 2014;42((12)):7720–7733. doi: 10.1093/nar/gku421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sharma A, Alswillah T, Singh K, Chatterjee P, Willard B, Venere M, et al. USP14 regulates DNA damage repair by targeting RNF168-dependent ubiquitination. Autophagy. 2018;14((11)):1976–1990. doi: 10.1080/15548627.2018.1496877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shire K, Wong AI, Tatham MH, Anderson OF, Ripsman D, Gulstene S, et al. Identification of RNF168 as a PML nuclear body regulator. J Cell Sci. 2016;129((3)):580–591. doi: 10.1242/jcs.176446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zemirli N, Pourcelot M, Dogan N, Vazquez A, Arnoult D. The E3 ubiquitin ligase RNF121 is a positive regulator of NF-κB activation. Cell Commun Signal. 2014;12((1)):72. doi: 10.1186/s12964-014-0072-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Geng R, Tan X, Wu J, Pan Z, Yi M, Shi W, et al. RNF183 promotes proliferation and metastasis of colorectal cancer cells via activation of NF-κB-IL-8 axis. Cell Death Dis. 2017;8((8)):e2994. doi: 10.1038/cddis.2017.400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gomes LR, Menck CFM, Leandro GS. Autophagy roles in the modulation of DNA repair pathways. Int J Mol Sci. 2017;18((11)):2351. doi: 10.3390/ijms18112351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miyamoto S. Nuclear initiated NF-κB signaling NEMO and ATM take center stage. Cell Res. 2011;21((1)):116–130. doi: 10.1038/cr.2010.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kanayama M, Inoue M, Danzaki K, Hammer G, He YW, Shinohara ML. Autophagy enhances NFκB activity in specific tissue macrophages by sequestering A20 to boost antifungal immunity. Nat Commun. 2015;6((1)):5779. doi: 10.1038/ncomms6779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shi CS, Kehrl JH. TRAF6 and A20 regulate lysine 63-linked ubiquitination of Beclin-1 to control TLR4-induced autophagy. Sci Signal. 2010;3((123)):ra42. doi: 10.1126/scisignal.2000751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang Y, Zhu WG, Zhao Y. Autophagy substrate SQSTM1/p62 regulates chromatin ubiquitination during the DNA damage response. Autophagy. 2017;13((1)):212–3. doi: 10.1080/15548627.2016.1245262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang L, Howell MEA, Sparks-Wallace A, Hawkins C, Nicksic CA, Kohne C, et al. p62-mediated Selective autophagy endows virus-transformed cells with insusceptibility to DNA damage under oxidative stress. PLoS Pathog. 2019;15((4)):e1007541. doi: 10.1371/journal.ppat.1007541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li D, Wang L, Fan Y, Song L, Guo C, Zhu F, et al. Down-regulation of A20 mRNA expression in peripheral blood mononuclear cells from patients with systemic lupus erythematosus. J Clin Immunol. 2012;32((6)):1287–1291. doi: 10.1007/s10875-012-9764-2. [DOI] [PubMed] [Google Scholar]
- 35.Hammer GE, Turer EE, Taylor KE, Fang CJ, Advincula R, Oshima S, et al. Expression of A20 by dendritic cells preserves immune homeostasis and prevents colitis and spondyloarthritis. Nat Immunol. 2011;12((12)):1184–1193. doi: 10.1038/ni.2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li M, Shi X, Qian T, Li J, Tian Z, Ni B, et al. A20 overexpression alleviates pristine-induced lupus nephritis by inhibiting the NF-κB and NLRP3 inflammasome activation in macrophages of mice. Int J Clin Exp Med. 2015;8((10)):17430–40. [PMC free article] [PubMed] [Google Scholar]
- 37.Dai C, Deng Y, Quinlan A, Gaskin F, Tsao BP, Fu SM. Genetics of systemic lupus erythematosus immune responses and end organ resistance to damage. Curr Opin Immunol. 2014;31:87–96. doi: 10.1016/j.coi.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sun L, Zou LX, Han YC, Wu L, Chen T, Zhu DD, et al. A20 overexpression exerts protective effects on podocyte injury in lupus nephritis by downregulating UCH-L1. J Cell Physiol. 2019 Feb;234((9)):16191–16204. doi: 10.1002/jcp.28282. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.