Figure 1.
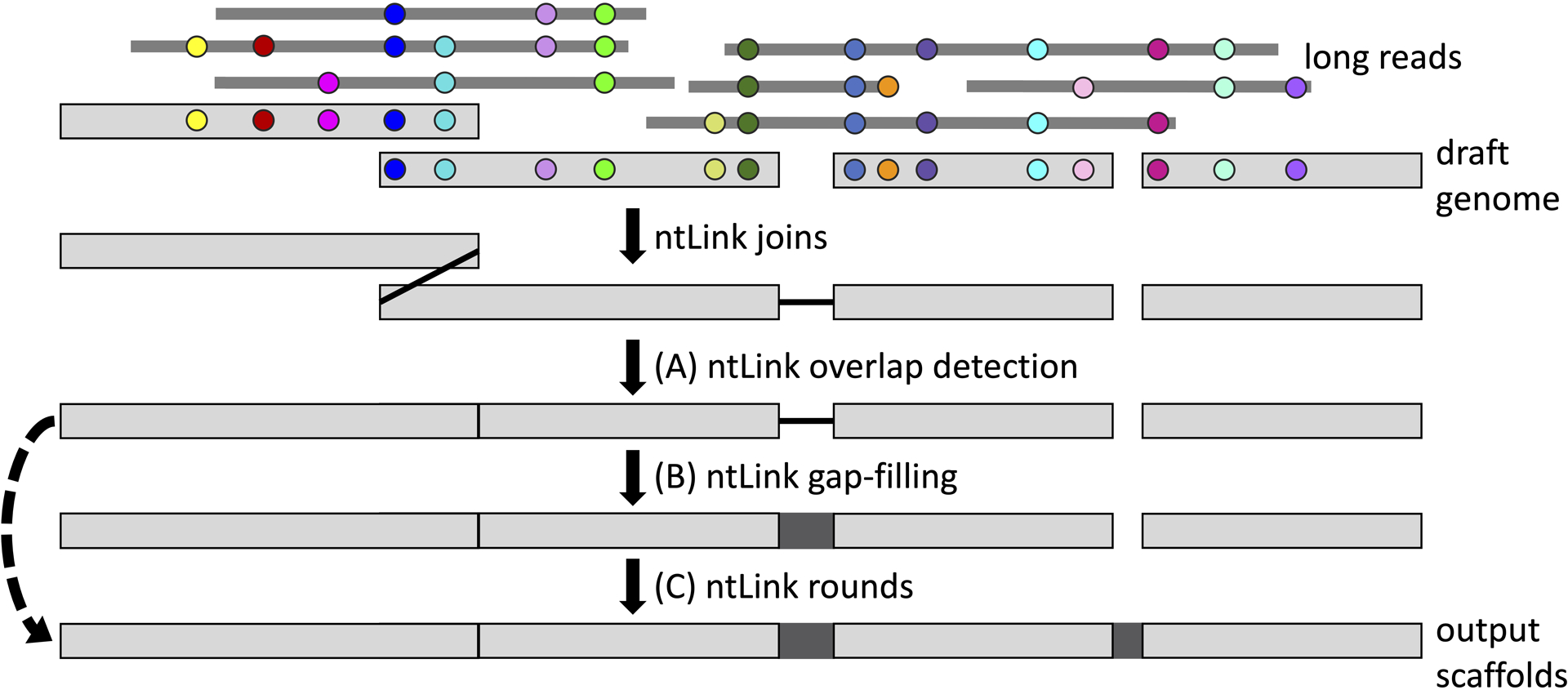
Flowchart showing the various available features for ntLink scaffolding. ntLink uses minimizers (indicated by coloured circles) to map the input long reads to the draft genome. Identical minimizers are represented by the same colour. ntLink uses these mappings to infer how the input contigs (draft genome sequences) should be ordered and oriented to produce genome scaffolds. (A) ntLink can also use minimizers to detect when adjacent contigs have an overlapping region, and resolves these overlaps (indicated by the vertical black line), as described in Basic Protocol 1. (B) In addition, as demonstrated in Basic Protocol 2, ntLink can use the input long-read information and minimizers to fill gaps between joined contigs (dark grey box). (C) Finally, as shown in Basic Protocol 3, running multiple in-code iterations of ntLink scaffolding can maximize the contiguity of the final output scaffolds. The ntLink rounds can be run with or without gap-filling, as indicated by the dashed transitive arrow.
