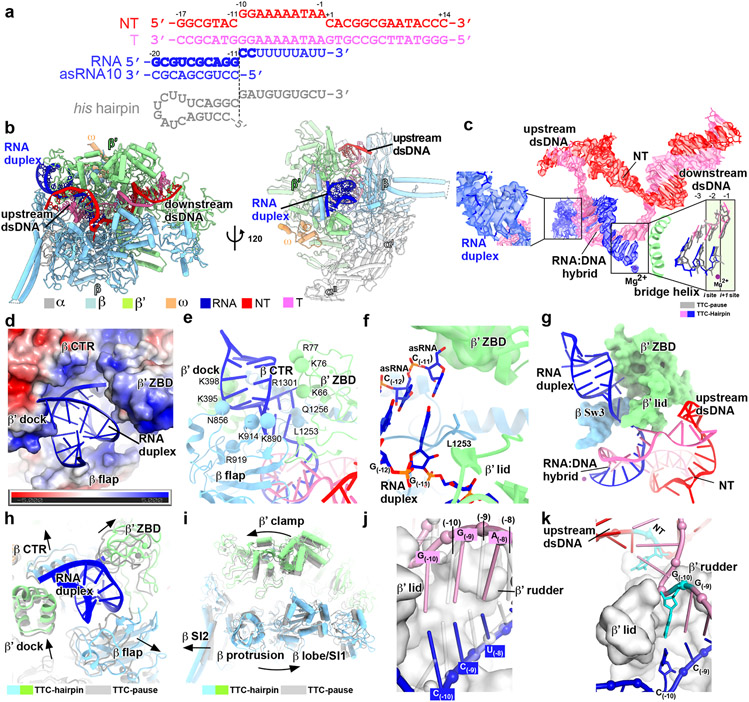
Figure 2. The TTC-hairpin complex.
a, The nucleic-acid scaffold and the antisense RNA (asRNA) used for cryo-EM structure determination. The potential hairpin-forming nucleotides of the nascent RNA are highlighted in bold. The dashed line defines the downstream edge of the RNA duplex in TTC-hairpin. The his hairpin is shown as a comparison. b, The structural model of TTC-hairpin in front and side views. c, The cryo-EM map for nucleic-acid scaffold. The left insert shows the map for the RNA duplex in the RNA exit channel and the right insert shows the superimposition of the base pairs (−3 to 1) of the RNA–DNA hybrid between TTC-hairpin (colored as in the scheme) and TTC-pause (gray). d, RNA duplex in the RNA exit channel. The electrostatic potential surface of RNAP was generated using APBS tools in Pymol. e, The detailed interaction of the RNA duplex with residues in the RNA exit channel. Spheres, the Cα atom of polar residues in H-bond distance with the phosphate backbone of RNA duplex. f, The interaction of the −11 base pair of RNA duplex with residues in the RNA exit channel. g, Further extension of RNA duplex is blocked by RNAP β′ ZBD, β′ lid, and β Sw3 motifs. h, The structural comparison of RNA exit channel between TTC-pause (gray) and TTC-hairpin (colored). i, The global conformational movement of TTC-hairpin (colored) compared with TTC-pause (gray). j, The comparison of the first three base pairs of the RNA–DNA hybrid between TTC-hairpin (pink and blue) and TEC (gray) (PDB: 6ALF). The two structures were superimposed based in the RNAP-β′ lid and rudder motifs. k, The G(−10) of the tDNA is under the tunnel formed by RNAP-β′ lid and rudder and ready to pair with the −10 nucleotide of ntDNA.