Abstract
Deep learning has emerged as the leading method in machine learning, spawning a rapidly growing field of academic research and commercial applications across medicine. Deep learning could have particular relevance to rheumatology if correctly utilized. The greatest benefits of deep learning methods are seen with unstructured data frequently found in rheumatology, such as images and text, where traditional machine learning methods have struggled to unlock the trove of information held within these data formats. The basis for this success comes from the ability of deep learning to learn the structure of the underlying data. It is no surprise that the first areas of medicine that have started to experience impact from deep learning heavily rely on interpreting visual data, such as triaging radiology workflows and computer‐assisted colonoscopy. Applications in rheumatology are beginning to emerge, with recent successes in areas as diverse as detecting joint erosions on plain radiography, predicting future rheumatoid arthritis disease activity, and identifying halo sign on temporal artery ultrasound. Given the important role deep learning methods are likely to play in the future of rheumatology, it is imperative that rheumatologists understand the methods and assumptions that underlie the deep learning algorithms in widespread use today, their limitations and the landscape of deep learning research that will inform algorithm development, and clinical decision support tools of the future. The best applications of deep learning in rheumatology must be informed by the clinical experience of rheumatologists, so that algorithms can be developed to tackle the most relevant clinical problems.
Introduction
Deep learning refers to a group of algorithms that use artificial neural networks and an optimization algorithm called backpropagation (with gradient descent) to model complex problems by learning complex functions that describe them (see Figure 1) (1). While deep learning methods have been designed and applied for many decades, it is only in the last 10 years that computer hardware has been able to train these increasingly complex models to such a level that they now dominate the machine learning landscape, both in terms of publications and performance. In recent years, the applications of deep learning in medicine have not only gained prominence but have started entering clinical practice. At the time of writing, the American College of Radiology lists 201 US Food and Drug Administration (FDA)–approved machine learning algorithms to support radiology (2), many of which use deep learning approaches. Deep learning methods power computers that beat grandmasters in Chess and Go (3), summarize documents as diverse as patents and academic papers (4), control autonomous cars (5), and predict and design macromolecules (6). Although still in its infancy, applications of deep learning in rheumatology are increasing across a broad range of areas (see Table 1). There are several ways to classify these various applications; however, one logical way to categorize deep learning algorithms is based on the input data type. Time‐series data are used for prediction tasks, written text is used for natural language processing, and images are used for computer vision. Here we explore rheumatic applications of deep learning across these 3 categories.
Figure 1.
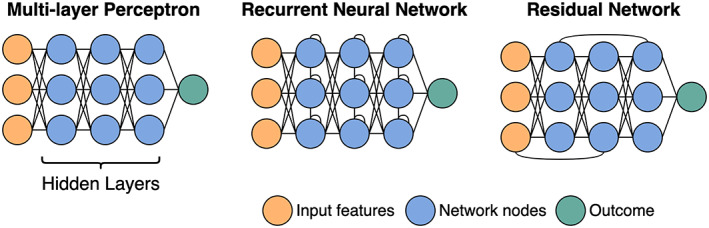
Neural network architectures. The first layer of a neural network consists of the data. These data are then passed to the first “hidden layer.” Each node, represented by a circle, is a weighted linear combination of all the nodes in the layer before. It is the weights that the model “learns.” Apart from a classic neural network where all nodes from 1 layer are connected to the next (otherwise known as a multilayer perceptron), other common architectures include recurrent networks with connections between nodes within a layer, usually used for sequence data (e.g., time‐series or text), and residual networks, where information from 1 layer can “skip” the next layer, giving the network a way to bypass inefficient layers.
Table 1.
Current applications of deep learning in rheumatology*
| Problem (source ref.) | Data type | Model | Implications |
|---|---|---|---|
| Identifying GCA features from temporal artery biopsy reports (13) | Text | Transformer | Accurate auditing of temporal artery biopsy reports can be performed using deep learning; however, this performance dropped when tested across centers. |
| Classifying HEp‐2 cells based on ANA IIF patterns (29) | Images | CNN | Automated ANA classification based on HEp‐2 cells is approaching expert human performance. |
| OESS from synovial ultrasound (44) | Images | CNN | Deep learning can identify synovitis on ultrasound with a high degree of accuracy. |
| SHS scoring using hand and foot radiographs (51) | Images | CNN | Radiographic scoring for RA is improving but still requires work for clinical implementation. |
| Predicting progression (any increase in K/L score) of knee OA based on baseline knee radiographs plus other clinical features (58) | Images | CNN | Radiographic progression in knee OA can be predicted with a combination of clinical features and baseline radiography using deep learning; however, there are unmeasured factors missing in these models. |
| Identifying halo sign on temporal artery ultrasound images (68) | Images | CNN | Deep learning has significant potential for automated identification of the halo sign; however, ensuring standardized image acquisition is a major barrier to implementation. |
| Predicting future RA disease activity (controlled versus uncontrolled) using clinical data from previous encounters (19) | EHRs | RNN | Deep learning can predict future disease activity from past disease activity and baseline factors; however, performance significantly dropped when the model was tested at a second center, suggesting that there is substantial heterogeneity between centers that must be accounted for in future models. |
GCA = giant cell arteritis; ANA = antinuclear antibody; IIF = immunofluorescence; CNN = convolutional neural network; OESS = EULAR Outcome Measures in Rheumatology synovitis scoring; SHS = modified Sharp/van der Heijde; RA = rheumatoid arthritis; K/L = Kellgren/Lawrence; OA = osteoarthritis; EHRs = electronic health records; RNN = recurrent neural network.
Learning from text
Natural language processing (NLP) is an interdisciplinary field of study with the main aim of having computers “perform useful tasks involving human language” (7). Traditionally, NLP has heavily relied on linguistic models of syntax and grammar, complemented by statistical analysis. In contrast, deep learning approaches in contemporary NLP rely less on assumptions about rules of natural language, including expertly curated words and phrases, building models capable of inferring those rules by learning from large bodies of text (8). Most recently, deep learning models for NLP have moved toward attention‐based models, in particular a group of models collectively referred to as “Transformers” (9). While previous state‐of‐the‐art NLP algorithms relied on modeling text as a sequence of words read one‐by‐one directionally (left‐to‐right for English text), Transformers allow the algorithm to view all the text at once and pick out the important words that provide context (see Figure 2). Attention‐based models, combined with pretraining on very large bodies of text (see section on transfer learning below), have allowed deep learning NLP algorithms to achieve state‐of‐the‐art performance on many language tasks.
Figure 2.
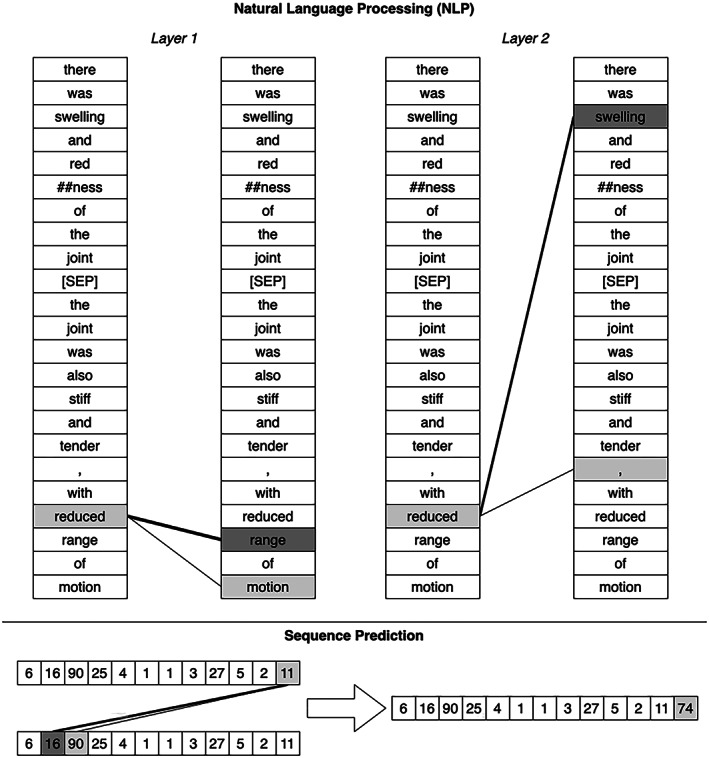
Visualization of attention model (ref. 94). Two attention layers are shown with text input for NLP (top). The original input text reads, “There was swelling and redness of the joint. The joint was also stiff and tender, with reduced range of motion.” This text is converted into tokens, sometimes splitting words into more than one token (here “redness” is split into “red” and “##ness”—the “##” signifying that this token belongs with the preceding token). On the left, a lower layer of the attention‐based model relied on the words “range” and “motion” to interpret the word “reduced.” On the right, at a higher layer, the word “reduced” also depends strongly on the word “swelling” in the previous sentence. An attention model can be used for any sequence data (bottom). Here, these numbers could be laboratory values, with the task of predicting the next value in the sequence. The attention layer used the values “16” and “90” to predict the next value in the sequence. In this instance, attention is used to focus on a similar pattern to anticipate a future value.
Classifying temporal artery biopsy reports
Classic NLP techniques have successfully been used for identifying patients with rheumatic diseases using electronic health records (EHRs) (10, 11, 12). These methods generally rely on the cultivation of a set of words and phrases strongly associated with the disease of interest. Given the wide phenotypic variability of rheumatic diseases, accurate identification and classification of patients based on clinical notes lends itself to deep learning techniques.
Presently, deep learning on text has only been applied in a single conference abstract, using Transformer models to classify temporal artery biopsy reports based on the presence of 3 histopathologic features (adventitial inflammation, giant cells, intimal hyperplasia) and overall conclusion (giant cell arteritis [GCA] or not) (13). This study used a model called DistilBERT (14), training on 161 biopsy reports from 1 center and testing both within that center and on 220 biopsy reports from a second center. The authors achieved excellent performance, particularly on the report conclusion (area under receiver operating characteristic curve [AUC] 0.99) and the presence of giant cells (AUC 0.99), with performance reducing slightly in the second center (AUC 0.93 and 0.97, respectively). Pooling data between the 2 centers and training a new model resulted in significant improvements (AUC 0.99 and 0.99, respectively) when tested on reports across both sites, suggesting that a diverse data set drawing on the language of multiple institutions will result in more generalizable models. It remains to be seen whether this model can generalize beyond 2 centers, particularly given the fact that both were within the same city in Australia—it is likely that variability in documentation practices, vocabulary, and idiomatic expressions results in reduced performance.
Learning from EHRs
Deep learning algorithms used for predicting future events are varied in design, but often rely on the use of time‐series data modeled as sequences. In this respect, predictive algorithms often use similar architectures to those used when learning from texts, which are also modeled as sequences. Many EHR prediction algorithms have been developed, most notably and commonly for inpatient outcomes such as length of stay and inpatient mortality rate (15). The nature of EHR data produces unique challenges, reflecting the bias of clinical decisions as much as patient physiology. Which data are missing and the presence of noise often reflects systematic decision making, rather than a random process (e.g., the absence of invasive blood pressure data in a critically ill patient may reflect a decision about the goals of care, rather than the lack of critical illness). Additionally, care must be taken to ensure data set leakage (16) does not occur, where the training data set is inadvertently informed by the testing data set (e.g., the same patient appears in both, but for different inpatient visits).
Predicting future diagnoses using EHRs
The Transformer architecture that has been successfully applied to NLP has demonstrated similar success in sequence models. Such models may use time‐sequence data to predict future events, drawing on the power of self‐attention, efficiently learning to recognize long‐range relationships between past events and future events (9). Li et al developed a Transformer model trained on 1.6 million primary care patients with at least 5 EHR encounters (17). For each individual, they created a sequence of EHR encounters, with the diagnoses and patient age at the time of the encounter forming the components of the sequence. The authors assessed predictive performance for a number of diseases, including rheumatic diseases. Most notably, the model was able to predict the future development of polymyalgia rheumatica (PMR) with very high accuracy (AUC 0.96). This result may partially reflect data quality issues, with PMR diagnosis in primary care frequently occurring without exclusion of alternative diagnoses (18); thus, the model is likely predicting the onset of a polymyalgic syndrome, rather than the specific diagnosis of PMR. Additionally, this algorithm may simply be identifying a pattern of clinical coding, rather than a sequence of clinical events—any model that uses clinical interpretation rather than patient physiology is prone to modeling not just patient outcomes, but also physician behavior.
Predicting disease activity using EHRs
Rheumatology, as a specialty dealing with chronic, relapsing–remitting disease, has maintained a strong interest in the task of predicting future disease activity. Frequent outpatient follow‐up with clinical and laboratory testing is used to detect changes in disease state and allow for interventions to treat any disease relapse or deterioration; yet precisely predicting who will experience relapse or deterioration remains a difficult task. Apart from the Transformer architecture mentioned above, for a long time, deep learning has approached the analysis of sequence data and prediction using recurrent neural networks (RNNs).
Norgeot et al (19) developed a model to predict whether patients with rheumatoid arthritis (RA) would have controlled or uncontrolled disease at their next clinic visit, based on data from the EHR. Their definition of disease control was based on a threshold cutoff of the Clinical Disease Activity Index (CDAI) (20). The input data included baseline measures (demographic characteristics, rheumatoid factor, citrullinated peptide antibody) and time‐dependent variables (laboratory values, medication, CDAI) from each visit. The time‐dependent variables were used to train an RNN—designed to identify periodicity and trend—to account for long‐range influences that might affect future disease states. The model was trained and tested on data from one hospital, before being further assessed on data from a different hospital. Predictably, the performance on data from the second hospital was inferior; however, the authors were able to improve the performance with a small amount of training on data from the new hospital, in a process known as transfer learning.
Learning from images
Computer vision is a field of image processing interested in automating tasks of visual perception. Since the groundbreaking AlexNet architecture won the ImageNet competition in 2012 (21), computer vision has been dominated by deep learning algorithms. The basis for its success to date has been one specific neural network architecture: the convolutional neural network (CNN). The CNN has had several inventors without reference to each other with slight variations; however, the precursor to modern CNN models is most frequently attributed to a 1999 paper by LeCun et al on object detection (22).
The building block of CNNs is a convolution kernel, a grid or matrix of numbers. The convolution passes over an image (a grid of numbers representing the individual pixels of an image), transforming the image in particular ways. Hard‐coded convolutions, like the one in Figure 3, may perform tasks like vertical edge detection. While convolutions have existed as an image processing technique for many decades, the innovation in deep learning is that the convolutions are not hard coded, they are learned. The layering of many convolutions allows a model to progressively build complex features. While early layers might learn convolutions for simple tasks like edge detection, later layers may join different edge detectors together to make object detectors, face detectors, and eventually solve complex tasks like facial expression detection.
Figure 3.
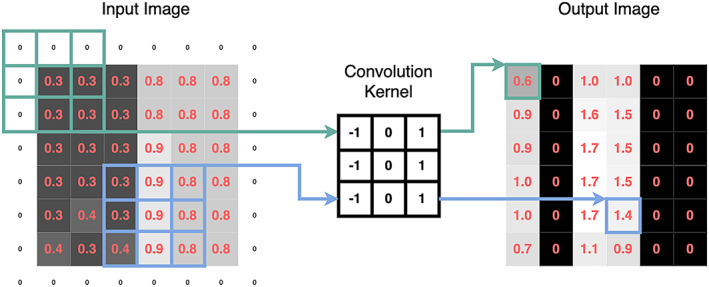
A vertical edge detector convolution kernel. Edges that transition from dark to light (as shown in the input image) will be light in the output image. The pixel values (representing light intensity) are shown as pink numbers. No values in the output image are <0. This is because all values <0 are turned into 0 by a function known as a rectified linear unit (ref. 95)—this is known as an activation function and is a common technique in deep learning. The rim of zeroes around the input image—known as “padding”—allows the output image to retain the same dimensions as the input image. Color figure can be viewed in the online issue, which is available at http://onlinelibrary.wiley.com/doi/10.1002/art.42296/abstract.
HEp‐2 image classification
Testing for antinuclear antibodies (ANAs) using indirect immunofluorescence (IIF) assays has been a cornerstone of the diagnostic evaluation of systemic autoimmunity for many decades (23). These methods rely on the visual inspection of HEp‐2 cells, mixed with fluorescence‐labeled antibodies from patient sera. Because different antigens are distributed differently within the HEp‐2 cells, the staining pattern produced by the fluorescence‐labeled antibodies can provide important diagnostic information about the antigen target and associated disease (24).
Automated analysis of HEp‐2 images has arisen as a field of research interest, motivated by concerns that the visual assessment of IIF patterns is subjective and time consuming (25). Recently, deep learning techniques have been applied to this task, with increasing success. Broadly, these techniques attempt to classify either individual HEp‐2 cells or specimens (containing many cells), by applying deep learning to coarsely labeled images. Rahman et al reviewed the application of deep learning techniques to these tasks, identifying 24 published methods for the classification of individual HEp‐2 cells and 7 methods for the classification of specimens (26).
A wide variety of deep learning techniques have been applied to cell classification. Broadly, deep learning has been applied in 2 ways to this task by 1) automatically extracting features and classifying cells or 2) automatically extracting features, which are then passed to an alternative model for classification. State‐of‐the‐art models using either technique have achieved accuracies exceeding 97% (26, 27), which favorably compare to human accuracy (73.3%) (28). However, this comparison has been criticized, as the task of classifying a single HEp‐2 cell, isolated from the broader context of the specimen, is not representative of how IIF tests for ANAs are performed in real clinical practice (29). Moreover, these methods are developed, tested, and validated using limited data sets (30, 31, 32, 33). These data sets lack consistency, both in terms of whether the images contain single cells or specimens, and the number of different staining categories classified.
In response to these data issues, Wu et al (29) curated a large data set of 51,694 HEp‐2 cell slides that more closely reflect clinical practice. These slides contain multiple cells per image, with up to 4 different patterns present in a single image. They tested multiple CNN architectures, ultimately finding that the Inception‐ResNet v2 architecture (34) had the best performance. In their testing data set, they found interobserver agreement—as measured using Cohen's kappa coefficient (35)—was similar between expert readers (0.85) and between expert readers and the final model (0.84).
Synovial ultrasound
Synovial ultrasound is an important and emerging technology in the diagnosis (36) and assessment (37) of inflammatory arthritis. Recently, the EULAR Outcome Measures in Rheumatology (EULAR–OMERACT) ultrasound taskforce developed a scoring system, sometimes referred to as the EULAR–OMERACT Synovitis Scoring (OESS) system, designed to be used as an outcome measure in clinical trials (37). While there is ongoing effort to validate and refine its use (38), the standardized application of an ultrasound synovitis scoring system is amenable to deep learning methods. Andersen et al (39) first applied 2 well‐studied CNN architectures to this problem (VGG‐16 [40] and Inception v3 [41]), after first performing pretraining on the popular ImageNet data set (42). They found overall good performance (AUC 0.93) for the task of discriminating healthy joints (OESS score 0–1) from unhealthy joints (OESS score 2–3); however, precise scoring on the ordinal scale did not appear to match human performance (43). A follow‐up method from the same group, this time using a so‐called “cascade” CNN (Figure 4A), showed similar performance compared to expert rheumatologists (44). In this algorithm, a CNN is given the task of classifying a power Doppler ultrasound image as either being at or above a given OESS grade. Each image that is classified as being of a higher grade is then passed to the next CNN, which performs an identical task for the next highest grade. These algorithms may be further enhanced with larger, multicenter data sets and further refinement of the CNN architecture. As in many medical applications, the laborious task of manually labeling images could be augmented by semisupervised learning and potentially by synthetic data from methods such as generative adversarial networks (45).
Figure 4.
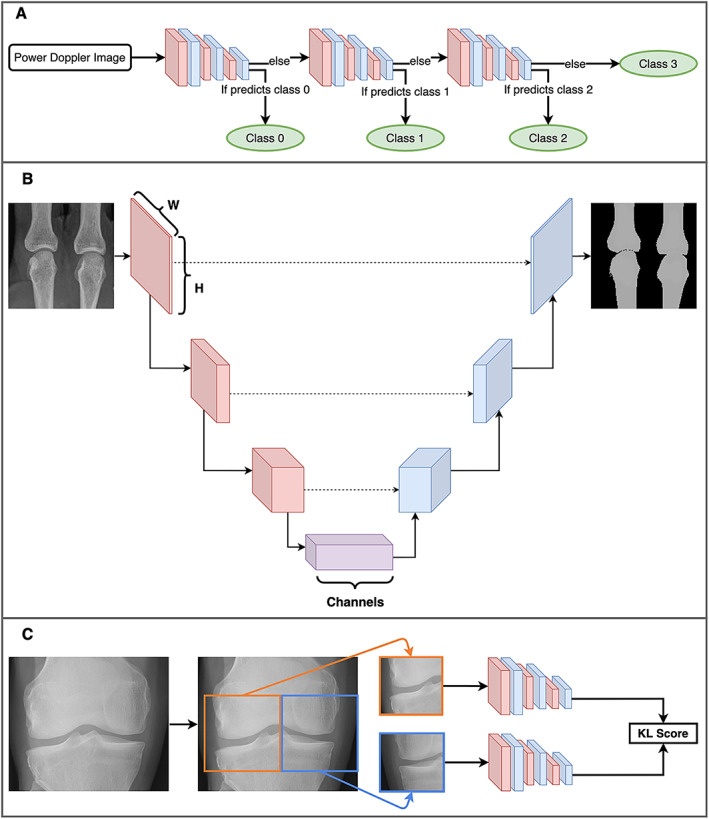
Three unique deep learning methods used in rheumatology. A, A cascade of convolutional neural networks (CNNs) used to classify power Doppler images. At each step, the CNN classifies the image as either a certain EULAR Outcome Measures in Rheumatology synovitis scoring class or any higher class (e.g., the first step classifies to either a class of 0 or >0). If the CNN determines that it belongs to a higher class, it is passed along to the next CNN, which performs the same task for the next highest class. Eventually, the final CNN simply classifies images as either class 2 or class 3. B, A simplified diagram of the U‐Net architecture (49). An image begins as an “N × N × C” shape, where “N × N” is the image size (e.g., 224 × 224 pixels) and “C” is the number of channels (typically 3 channels of red/green/blue for a color image). The model gradually reduces the size, while increasing the number of channels, until the bottom of the architecture is reached, and then the reverse occurs. Connections across the architecture (dashed lines) act as a “memory.” The image recovered at the end is a segmented image, partitioning the original into the relevant parts. In this example, the bones of 2 metacarpal joints are segmented from the plain radiograph. C, A single coronal radiograph of the knee joint split into 2 images: the right half of the knee and the horizontally flipped left half. Both images are passed through the same CNN before joining up to produce a Kellgren/Lawrence (K/L) composite score as the model output. Color figure can be viewed in the online issue, which is available at http://onlinelibrary.wiley.com/doi/10.1002/art.42296/abstract.
Joint damage in inflammatory arthritis
Progressive joint erosion early in the course of RA is one of the major predictors of future physical function (46). The prevention of progressive joint erosions is an important outcome measure in establishing the efficacy of any disease‐modifying agent in RA and other inflammatory arthritides. It is therefore important that erosive disease is measured consistently and with high sensitivity. The modified total van der Heijde Sharp Score (mTSS) (47) is commonly used in clinical trials to assess progressive joint erosions, consisting of both an erosion score and joint space narrowing score (JSN). The process of grading these components is a visual task performed by trained radiologists and rheumatologists, making it a good application for neural network models.
Hirano et al developed a neural network model to grade hand joints according to mTSS scores using hand radiographs (48). To perform joint‐level scoring, their model first had to perform a task called image segmentation to create bounding boxes around individual joints so they could be assessed. Rather than utilizing a deep learning model for this task, such as the popular U‐Net model (Figure 4B) first developed for biomedical image segmentation (49), the authors used hard‐coded convolutions known as Haar‐like features, in a method first described by Viola et al (50). After image segmentation, the authors built JSN and erosion score models with 2 convolutional layers and 3 fully connected layers. The JSN and erosion score models had similar performance compared to clinician assessment (correlation coefficients 0.72–0.88 and 0.54–0.75, respectively); however, overall sensitivity to detect erosions was low (34.8–42.4%). The major limitation of this algorithm was a relatively small data set (186 radiographs). With larger data sets, deeper models with more sophisticated architectures will perhaps make this a clinically applicable scenario for deep learning.
Recently, deep learning methods using a combination of CNNs and attention mechanisms (51) have been used for mTSS scoring in RA. The authors also used a 2‐stage approach, first using a CNN architecture called RetinaNet (52) to detect joint groups, followed by another CNN architecture called EfficientNet (53) to score individual joints. Additionally, the authors applied an attention layer (Figure 2) after the convolutional layers. The attention layer effectively constrains the area of interest to only those pixels in the image that provide a substantial contribution to the final prediction—the attention to all other pixels becomes negligible. By interrogating this attention layer, the authors generated heatmaps to demonstrate which regions contribute to the scoring of a joint, although interpreting these as an explanation of how the model works should be treated with great caution (see section on explainability below) (54, 55).
Plain films in knee osteoarthritis (OA)
Current EULAR recommendations for the diagnosis of OA only support imaging as a diagnostic tool in atypical presentations of suspected OA (56); however, this recommendation is not supported by high‐level evidence and the role of routine imaging remains a topic of debate (57). Outside of diagnosis, there is also debate about the prognostic role of imaging features to predict symptom severity and progression. The current EULAR recommendations do not support the use of imaging for prognostication; however, this is on the basis of older studies using hand‐crafted imaging features, not the systematic discovery of prognostic features from deep learning algorithms.
Tiulpin et al developed deep learning models for diagnosis and prognosis of knee OA using plain radiographs (58, 59, 60, 61). For diagnosis, the authors utilized CNN architectures to train 2 models to grade images according to OA severity. In their first model, they used a Siamese network to grade knee radiographs according to the Kellgren/Lawrence (K/L) composite score (62), a global rating system for knee OA (scale 0–4). The Siamese Network, as first proposed by Baldi and Chauvin (63), trains 2 identical neural networks simultaneously, with the task of determining whether 2 images meet some similarity threshold—in the original paper, the models compare 2 fingerprints to determine whether they come from the same finger. In the present study, the input image pairs were automatically segmented from the original plain radiographs, consisting of the right half of the tibiofemoral joint and the horizontally flipped left half. Because the tibiofemoral joint has horizontal symmetry with respect to the features that predict K/L score (Figure 4C), a single model can identify salient features from both sides (provided one half is horizontally flipped). These left‐ and right‐sided predictions are then joined to provide an overall K/L score. Overall, they found good agreement between model and clinician scores, with a Cohen's kappa coefficient of 0.83.
The second diagnostic model from Tiulpin et al used a conventional ResNet architecture with transfer learning from ImageNet to grade both individual OA features using the Osteoarthritis Research Society International (OARSI) atlas of OA radiographic features (64) and K/L score. The model was able to achieve state‐of‐the‐art results on OARSI scoring, exceeding human accuracy on this task.
For the task of prognostication, the authors trained a CNN on baseline knee radiographs to predict whether repeat radiography would show an increase in K/L score (58). They compared this model to a model that used tabular data: age, sex, body mass index, K/L grade, Western Ontario and McMaster Universities Osteoarthritis Index, injury, and surgery history. Additionally, they combined these 2 models to test whether there was any additional benefit from a so‐called “multimodal” approach. Their CNN model outperformed the tabular data model, while the multimodal approach outperformed the individual models, demonstrating that knee radiographs contain prognostic features not present in structured patient data, even reported K/L grade. Despite accurate radiographic scoring, applicability will ultimately be limited given the poor correlation between radiographic scoring and clinical outcomes such as pain scores (65). Predicting progressive disease may only be helpful if the definition of progressive disease is a clinical, rather than radiographic outcome.
Temporal artery ultrasound
The diagnosis of GCA is a vexing problem for rheumatologists, in no small part due to the lack of an accurate noninvasive diagnostic test. Temporal artery ultrasound is one emerging solution to this problem, with EULAR guidelines recommending ultrasound as the first‐line imaging modality for the evaluation of suspected GCA (66). Despite its relatively good performance as a diagnostic test, temporal artery ultrasound suffers from only moderate interrater agreement, with significant training requirements that pose a barrier to the widespread adoption of this test (67). Deep learning therefore appears attractive as a tool to reduce the variability in interpretation and perhaps even lower the barrier to the adoption of this test.
Roncato et al (68) developed a CNN model, specifically to identify 1 common feature in positive temporal artery ultrasounds: the halo sign (69). The first step in their algorithm was to perform semantic segmentation of transverse and longitudinal color Doppler or power Doppler images of temporal arteries. This process involved drawing boxes around arteries and adjacent tissue, with individual pixels in these boxes labeled as either halo sign–positive or halo sign–negative. The authors used a U‐Net model, designed specifically for biomedical image segmentation (49). The final classification of each image as either positive or negative was based on the percentage of pixels within the bounding box classified as halo sign–positive—a higher percentage of halo sign pixels means a higher probability that the image truly contains a halo sign. The accuracy of the model was compared using 2 groups of images: group 1 obtained by a single operator, using a standardized protocol; and group 2 obtained by multiple different operators using a variety of parameters. The performance on group 1 was significantly higher than group 2 (AUC 0.95 versus 0.82). Although training on more nonstandard images may improve performance, deep learning can also be used for computer‐assisted image acquisition to assist clinicians and sonographers in acquiring standardized views at the time of ultrasound.
The future
Learning with limited data
One of the main features of deep learning is the ability to capitalize on the wealth of large data sets, with improvements in computer vision models seen even beyond massive data sets composed of 300 million images (70). Despite this, there are 3 established and emerging technical solutions to the problem of learning with limited data (See Figure 5).
Figure 5.
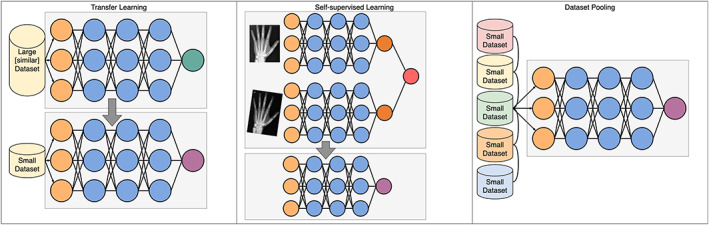
Three methods to overcome the complications of limited data sets. Transfer learning takes a model trained on a large data set and repurposes it for a new task, replacing only the final layer. Self‐supervised learning is a type of transfer learning; however, the data set used in pre‐training does not need to have labels—here the task is simply to recognize that 2 versions of the same image are indeed the same image, and in doing so the model learns to recognize invariant features. Increasing data set size can be done in a number of ways; however, pooling data across institutions has technical, logistical, and privacy issues that must be overcome. Circles represent individual nodes. Color figure can be viewed in the online issue, which is available at http://onlinelibrary.wiley.com/doi/10.1002/art.42296/abstract.
Transfer learning
The first method, now well‐established in deep learning research and applications, is a concept known as transfer learning. Transfer learning is the process whereby a model is trained on a large data set (pretraining), and then only partially retrained on a small data set, even from a different domain (e.g., a model trained on photographs is retrained on radiographs). The motivation for this process is that, in training a large data set, the model will have learned generalizable properties. These generalizable properties are believed to be learned in the early layers, and so it is only the final layers that are retrained on the new, smaller data set of interest. This has the effect of “specializing” a pretrained model for a downstream task. The pretraining process is generally performed using a process called self‐supervised learning, which does not require any hand‐labeled data, but instead learns by predicting missing words, the next word in a sentence or similar tasks. In certain circumstances, these pretrained models can be used in a few‐ or zero‐shot setting, meaning they are fine‐tuned on few or even no examples of the downstream task. This is particularly the case for NLP, where the task can be distinguished by a text prompt (e.g., a clinical question)—if the pretrained model can interpret the prompt, then it may not necessarily need to see any examples in order to perform the task.
Self‐supervised learning
Recently, self‐supervised learning has emerged as a method to learn from large, unlabeled data sets (71). Self‐supervision is achieved by creating tasks that allow models to learn generalizable features. For example, in NLP, learning to predict the next word in a sentence requires learning common linguistic features, such as sentence structure and word meaning. In computer vision, learning to pair original and distorted versions of the same image requires learning invariant features, such as the rounded shape of the metacarpal head. Self‐supervised models can then be used in a transfer learning process to train on smaller, labeled data sets. Given the relatively small data sets in rheumatology, it is highly likely that new applications for deep learning will be powered by self‐supervised learning and transfer learning.
Methods of increasing data set size
While single institutions might generate only relatively small data sets, pooling data across many institutions can result in large data sets. Large data sets may be necessary, particularly where outcomes are rare. In rheumatology, the Rheumatology Informatics System for Effectiveness registry has begun pooling EHR data to further our understanding of rheumatic diseases (72); however, further barriers exist where text and image data are needed. Concerns about data privacy can be a barrier to sharing data across institutional, regional, and international borders—aside from specific data sharing agreements, much effort has been made in producing technical solutions to this problem.
Recently, federated learning has emerged as a powerful tool to allow multiple sites to contribute training data without ever sharing raw data (73). Several federated learning techniques exist; however, recent work has focused on methods whereby individual sites train small models on local data, with the coefficients of these models sent to a central site that uses these to train a multicenter model (74, 75). Federated learning is not without significant challenges, including the high cost of setting up a central server and communication between sites, black box models (particularly if only model weights are shared and not raw data), and no clear best way to aggregate data from heterogeneous sites (76). In addition to these concerns, federated learning does not fully solve the data privacy problem; all models can “memorize” training data and therefore if privacy is a significant concern, models have to be trained using a special technique called differential privacy to prevent data memorization (77).
When there is not even enough data to pool, or data pooling is impractical, an alternative practice is to generate synthetic data. Generating artificial samples has been an area of intense research in deep learning, particularly after the development of Generative Adversarial Networks (GANs) (45) by Goodfellow et al in 2014. GANs train 2 networks simultaneously: a generator network to generate new data and a discriminator network to discriminate real data from synthetic data. Although these models can be difficult to train, as it becomes harder and harder for the discriminator to distinguish between real and synthetic data, the quality of synthetic data increases.
Data integrity, bias, and ethics
Although deep learning has a long history in computer science research, it has only been the relatively recent development of computers capable of training these algorithms that has resulted in an explosion of applications in medicine. In rheumatology, these methods have only now begun to show promise—although not without potential impediments. As deep learning methods gain more prominence, issues of data integrity and standardization will become more important. As we saw above, the standardized acquisition of temporal artery ultrasound images poses a potential barrier to the effective deployment of deep learning algorithms to diagnose GCA. Outside of rheumatology, a similar problem in echocardiography has led to several FDA‐approved ultrasound machines that not only interpret images, but guide the user on how to adjust the probe to obtain enhanced views (78). As rheumatologists and machine learning engineers begin applying deep learning techniques to clinical problems, we will need an even greater focus on data integrity and quality. Beyond simply ensuring high quality data, careful collection and curation of data sets is required when considering how data may reflect the systematic biases against marginalized and underrepresented minorities in our communities, making resultant algorithms unsafe and inaccurate for many.
New guidelines for reporting clinical trials of artificial intelligence interventions provide a guide for the steps required to demonstrate safety and efficacy of these algorithms (79). Like clinical trials of conventional medical interventions, careful trial design is paramount in testing clinical algorithms. Unlike conventional trials, the risk of bias extends beyond the study design and into the algorithm design itself. The data set or sets used to train deep learning algorithms can introduce substantial bias that may not only invalidate the results, but also introduce racial, sex‐based, or other forms of discrimination if applied systematically. Promising methods to identify and minimize such bias include report cards for evaluating performance in particular groups (80); however, static model checks are not enough in production, and the predictions of any model should be periodically examined to ensure they are not introducing or perpetuating unacceptable bias. For more detail on bias in clinical machine learning, Chen et al (81) provide a detailed overview of how bias is introduced at each step of the algorithm development process, while Gianfrancesco et al (82) detail the sources of bias in EHR‐based models.
New technologies like wearable devices, assisted by automatic deep learning algorithms, have the potential to diagnose a myriad of conditions under circumstances never seen before at such scale (e.g., asymptomatic atrial fibrillation in young people), with the resulting risk of widespread overdiagnosis. In rheumatology, for example, the unsolved problem of treatment for clinically suspect arthralgia may be compounded should automated diagnostic tools become available, identifying subclinical reductions in morning mobility leading to a tidal wave of very early arthritis diagnoses. While coordinated data collection will be needed to quantify the risks associated with these new diagnostic paradigms (83), this also presents a new opportunity to further our understanding of rheumatic diseases by studying longitudinal cohorts from very early disease stages. Compounding this, access to these devices is contingent on affordability (84)—one way in which technologically enhanced medicine widens the socioeconomic disparities in the provision of health care.
Explainability in rheumatology
Definition of explainability
Neural networks are often described as black boxes, in that their internal processes are inscrutable to humans. Many have argued that we need ways to explain why a model reaches a decision so that doctors are able to interpret it and apply it clinically (85). The most common methods for explainability in conventional machine learning—in particular Shapley Additive Explanations and Local Interpretable Model‐Agnostic Explanations (86)—are broadly classified as post hoc perturbation methods. These algorithms perturb the input data and measure how these perturbations in the input alter the model output. Although post hoc explainability methods have shortcomings, including susceptibility to hide model bias (86), even inadequate model explanations provide interpretable outputs when the input variables are themselves simple and interpretable (e.g., how does cardiovascular risk change if the patient has hypertension). Compared to conventional machine learning methods, explainability methods for deep learning are more difficult to interpret. In medical imaging, the most common method of explainability is saliency maps, which visually show the parts of the input image that most contributed to the final prediction (87).
Strengths
Explainability methods may have a role in evaluating models. As previously discussed, a 2019 article predicted future Clinical Disease Activity Index (CDAI) and used permutation importance scores to determine feature importance. The authors concluded that disease activity, laboratory test values, and medications were the best predictors of future CDAI (19). With simple input data, this method provides useful information regarding how the model operates, but when dealing with complex data (e.g., text, images), interpretation is unclear.
Weaknesses
In a model diagnosing knee OA, saliency maps were used to show that the model was identifying relevant radiologic features (60). The authors found that osteophytes were highlighted and concluded that attention maps would “build better trust in the clinical community.” However, they also acknowledge that the reason these anatomically relevant areas were highlighted was because they constrained the model to only assess these regions. Additionally, a model developed to predict RA radiographic scores used saliency maps to determine which joints were predictive of the scores (51). While these images show some focus over bone and joint space, they also highlight parts of the image that are empty.
In both of these cases, the meaning of these explanations is unclear. In the first, it was inevitable that these regions would be highlighted, while in the second case, the model uses areas that have no anatomical relevance in its prediction. Both papers use this as evidence that their model is performing reliably. Even more problematically, it has been shown that in models where the input image is modified to result in an incorrect prediction, the saliency map can still highlight clinically relevant regions of the image (88). This is falsely reassuring that the model is behaving appropriately, while still providing the wrong answer. A recent paper highlighted these concerns about the danger of relying on such methods to engender trust in a system and suggests we ought to depend on rigorous evaluation instead (54). Whether or not saliency maps produce sensible explanations, they should not be what we rely on to trust the behavior of neural networks.
Evaluation
Although explainability techniques can tell us something about model behavior, they cannot tell us how to interpret the predictions. Three steps are need for safe implementation: testing on external data sets is needed to ensure models generalize to different population, performance should be evaluated in subgroups to eliminate bias (89), and, ultimately, models should be tested in randomized control trials. It is vital to understand how models affect patient outcomes in clinical settings and it is this, not explainability techniques, that rheumatologists should be demanding before a model reaches clinical implementation.
Translation into practice
Deep learning is transforming many industries and at an ever‐increasing pace. For example, Google's language translation (90), Uber's expected arrival time (ETA) prediction (91), and Microsoft's code completion tool (92) are all deep learning algorithms that many people rely on daily. Uber switched their ETA algorithm to deep learning because of its ability to easily scale up with larger data sets and larger models. However, the same cost/benefit tradeoff is not always clear in medicine, where the scale may not be so large and financial barriers, such as the cost of regulatory approval, may be a substantial impediment. The barriers to widespread adoption are necessarily set high, with minimum standards of safety and efficacy set not only by the regulatory authorities, but also the clinicians who must “buy in” to this new technology. Nevertheless, the deep learning revolution has been largely driven by falling costs in computing power (93), with no clear indication that this will significantly plateau. We can therefore expect further improvements, shifting the cost/benefit tradeoff and encouraging even greater investment in research. Who takes advantage of this, whether it be academic or industry, is an open question.
Conclusion
Deep learning is an important method in medical machine learning applications and will likely become the dominant method in the future. Several applications of deep learning in rheumatology have been reported, with the promise of many more to come. Importantly, although deep learning methods offer the opportunity to improve the efficiency of some clinical tasks, they also provide a powerful technique for generating new knowledge and insights, particularly in the previously impenetrable analysis of unstructured data such as text and images. To date, much of the published work applying deep learning in rheumatology has occurred on small, homogeneous public data sets that do not reflect the diversity of real data, including the interactions between different data modalities (e.g., EHRs plus imaging). Further collaboration and interaction between machine learning researchers and rheumatology researchers will likely result in more clinically applicable algorithms, with translation into clinical practice being the next great hurdle to overcome. Researchers should therefore be familiar with the potential applications and limitations of these methods in their own research, and clinicians should be familiar with some of the potential benefits and pitfalls as these methods make their way into clinical practice.
AUTHOR CONTRIBUTIONS
All authors were involved in drafting the article or revising it critically for important intellectual content, and all authors approved the final version to be published.
Supporting information
Disclosure Form
ACKNOWLEDGMENT
Open access publishing was facilitated by The University of Melbourne, as part of the Wiley ‐ The University of Melbourne agreement via the Council of Australian University Librarians.
Author disclosures are available at https://onlinelibrary.wiley.com/action/downloadSupplement?doi=10.1002%2Fart.42296&file=art42296‐sup‐0001‐Disclosureform.pdf.
REFERENCES
- 1. LeCun Y, Bengio Y, Hinton G. Deep learning. Nature 2015;521:436–44. [DOI] [PubMed] [Google Scholar]
- 2. American College of Radiology . AI Central. URL: https://aicentral.acrdsi.org/.
- 3. Silver D, Hubert T, Schrittwieser J, et al. Mastering chess and shogi by self‐play with a general reinforcement learning algorithm. arXiv [cs.AI] 2017. URL: http://arxiv.org/abs/1712.01815. [DOI] [PubMed]
- 4. Zhang J, Zhao Y, Saleh M, et al. PEGASUS: Pre‐training with Extracted Gap‐sentences for Abstractive Summarization. arXiv [cs.CL] 2019. URL: http://arxiv.org/abs/1912.08777.
- 5. Chen K, Oldja R, Smolyanskiy N, et al. MVLidarNet: Real‐time multi‐class scene understanding for autonomous driving using multiple views. arXiv [cs.CV] 2020. URL: http://arxiv.org/abs/2006.05518.
- 6. Jumper J, Evans R, Pritzel A, et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021;596:583–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jurafsky D, Martin JH. Speech and language processing: an introduction to natural language processing, computational linguistics, and speech recognition. Prentice Hall: Hoboken; 2009. [Google Scholar]
- 8. Young T, Hazarika D, Poria S, et al. Recent trends in deep learning based natural language processing [review]. IEEE Comput Intell Mag 2018;13:55–75. [Google Scholar]
- 9. Vaswani A, Shazeer N, Parmar N, et al. Attention is all you need. In: Guyon I, Luxburg UV, Bengio S, et al, editors. Advances in neural information processing systems 30. Curran Associates, Inc: Red Hook (New York); 2017. p. 5998–6008. [Google Scholar]
- 10. Liao KP, Cai T, Gainer V, et al. Electronic medical records for discovery research in rheumatoid arthritis. Arthritis Care Res (Hoboken) 2010;62:1120–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhao SS, Hong C, Cai T, et al. Incorporating natural language processing to improve classification of axial spondyloarthritis using electronic health records. Rheumatology (Oxford) 2020;59:1059–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tedeschi SK, Cai T, He Z, et al. Classifying pseudogout using machine learning approaches with electronic health record data. Arthritis Care Res (Hoboken) 2021;3:442–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. McMaster C, Yang V, Sutu B, et al. Temporal artery biopsy reports can be accurately classified by artificial intelligence [abstract]. Arthritis Rheumatol 2021;73 Suppl. URL: https://acrabstracts.org/abstract/temporal‐artery‐biopsy‐reports‐can‐be‐accurately‐classified‐by‐artificial‐intelligence/. [Google Scholar]
- 14. Sanh V, Debut L, Chaumond J, et al. DistilBERT, a distilled version of BERT: smaller, faster, cheaper and lighter. arXiv [cs.CL] 2019. URL: http://arxiv.org/abs/1910.01108.
- 15. Rajkomar A, Oren E, Chen K, et al. Scalable and accurate deep learning with electronic health records. NPJ Digit Med 2018;1:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Elangovan A, He J, Verspoor K. Memorization vs. generalization: quantifying data leakage in NLP performance evaluation. arXiv [cs.CL] 2021. URL: http://arxiv.org/abs/2102.01818.
- 17. Li Y, Rao S, Solares JR, et al. BEHRT: transformer for electronic health records. Sci Rep 2020;10:7155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Helliwell T, Hider SL, Mallen CD. Polymyalgia rheumatica: diagnosis, prescribing, and monitoring in general practice. Br J Gen Pract 2013;63:e361–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Norgeot B, Glicksberg BS, Trupin L, et al. Assessment of a deep learning model based on electronic health record data to forecast clinical outcomes in patients with rheumatoid arthritis. JAMA Netw Open 2019;2:e190606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Aletaha D, Nell VP, Stamm T, et al. Acute phase reactants add little to composite disease activity indices for rheumatoid arthritis: validation of a clinical activity score. Arthritis Res Ther 2005;7:R796–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Krizhevsky A, Sutskever I, Hinton GE. ImageNet classification with deep convolutional neural networks. In: Pereira F, Burges CJ, Bottou L, et al, editors. Advances in neural information processing systems 25. Curran Associates, Inc: New York; 2012. p. 1097–105. [Google Scholar]
- 22. LeCun Y, Haffner P, Bottou L, et al. Object recognition with gradient‐based learning. In: Forsyth DA, Mundy JL, di Gesú V, et al, editors. Shape, contour and grouping in computer vision. Springer: Berlin Heidelberg; 1999. p. 319–45. [Google Scholar]
- 23. Mahler M, Meroni PL, Bossuyt X, et al. Current concepts and future directions for the assessment of autoantibodies to cellular antigens referred to as anti‐nuclear antibodies. J Immunol Res 2014;2014:315179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Damoiseaux J, Andrade LE, Carballo OG, et al. Clinical relevance of HEp‐2 indirect immunofluorescent patterns: the International Consensus on ANA patterns (ICAP) perspective. Ann Rheum Dis 2019;78:879–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Infantino M, Meacci F, Grossi V, et al. The burden of the variability introduced by the HEp‐2 assay kit and the CAD system in ANA indirect immunofluorescence test. Immunol Res 2017;65:345–54. [DOI] [PubMed] [Google Scholar]
- 26. Rahman S, Wang L, Sun C, et al. Deep learning based HEp‐2 image classification: a comprehensive review. arXiv [cs.CV] 2019. URL: http://arxiv.org/abs/1911.08916. [DOI] [PubMed]
- 27. Vununu C, Lee SH, Kwon KR. A strictly unsupervised deep learning method for HEp‐2 cell image classification. Sensors 2020;20:2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Foggia P, Percannella G, Soda P, et al. Benchmarking HEp‐2 cells classification methods. IEEE Trans Med Imaging 2013;32:1878–89. [DOI] [PubMed] [Google Scholar]
- 29. Wu YD, Sheu RK, Chung CW, et al. Application of supervised machine learning to recognize competent level and mixed antinuclear antibody patterns based on ICAP International Consensus. Diagnostics (Basel) 2021;11:642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Benammar Elgaaied A, Cascio D, Bruno S, et al. Computer‐assisted classification patterns in autoimmune diagnostics: the AIDA Project. Biomed Res Int 2016;2016:2073076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Heo‐2 Benchmarking . Datasets & Tools: HEp‐2 contest @ ICPR 2016. URL: https://hep2.unisa.it/dbtools.html.
- 32. Mivia . ICPR 2012–Contest on HEp‐2 cells classification. URL: https://mivia.unisa.it/contest-hep-2/.
- 33. Wiliem A, Wong Y, Sanderson C, et al. Classification of human epithelial type 2 cell indirect immunofluoresence images via codebook based descriptors. URL: http://staff.itee.uq.edu.au/lovell/snphep2/.
- 34. Kassani SH, Kassani PH, Khazaeinezhad R, et al. Diabetic retinopathy classification using a modified Xception architecture. In: 2019 IEEE International Symposium on Signal Processing and Information Technology (ISSPIT); 2019. p. 1–6. [Google Scholar]
- 35. Cohen J. A coefficient of agreement for nominal scales. Educ Psychol Meas 1960;20:37–46. [Google Scholar]
- 36. Kaeley GS, Bakewell C, Deodhar A. The importance of ultrasound in identifying and differentiating patients with early inflammatory arthritis: a narrative review. Arthritis Res Ther 2020;22:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. D'Agostino MA, Terslev L, Aegerter P, et al. Scoring ultrasound synovitis in rheumatoid arthritis: a EULAR‐OMERACT ultrasound taskforce‐Part 1: definition and development of a standardised, consensus‐based scoring system. RMD Open 2017;3:e000428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Terslev L, Christensen R, Aga AB, et al. Assessing synovitis in the hands in patients with rheumatoid arthritis by ultrasound: an agreement study exploring the most inflammatory active side from two Norwegian trials. Arthritis Res Ther 2019;21:166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Andersen JK, Pedersen JS, Laursen MS, et al. Neural networks for automatic scoring of arthritis disease activity on ultrasound images. RMD Open 2019;5:e000891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Simonyan K, Zisserman A. Very deep convolutional networks for large‐scale image recognition. arXiv [cs.CV] 2014. URL: http://arxiv.org/abs/1409.1556.
- 41. Szegedy C, Vanhoucke V, Ioffe S, et al. Rethinking the inception architecture for computer vision. arXiv [cs.CV] 2015. URL: http://arxiv.org/abs/1512.00567.
- 42. Russakovsky O, Deng J, Su H, et al. ImageNet large scale visual recognition challenge. Int J Comput Vis 2015;115:211–52. [Google Scholar]
- 43. Terslev L, Naredo E, Aegerter P, et al. Scoring ultrasound synovitis in rheumatoid arthritis: a EULAR‐OMERACT ultrasound taskforce‐Part 2: reliability and application to multiple joints of a standardised consensus‐based scoring system. RMD Open 2017;3:e000427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Christensen AB, Just SA, Andersen JK, et al. Applying cascaded convolutional neural network design further enhances automatic scoring of arthritis disease activity on ultrasound images from rheumatoid arthritis patients. Ann Rheum Dis 2020;79:1189–93. [DOI] [PubMed] [Google Scholar]
- 45. Goodfellow IJ, Pouget‐Abadie J, Mirza M, et al. Generative adversarial networks. arXiv [stat.ML] 2014. URL: http://arxiv.org/abs/1406.2661.
- 46. Ødegård S, Landewé R, van der HD , et al. Association of early radiographic damage with impaired physical function in rheumatoid arthritis: a ten‐year, longitudinal observational study in 238 patients. Arthritis Rheum 2006;54:68–75. [DOI] [PubMed] [Google Scholar]
- 47. Van der Heijde D. How to read radiographs according to the Sharp/van der Heijde method. J Rheumatol 2000;27:261–3. [PubMed] [Google Scholar]
- 48. Hirano T, Nishide M, Nonaka N, et al. Development and validation of a deep‐learning model for scoring of radiographic finger joint destruction in rheumatoid arthritis. Rheumatol Adv Pract 2019;3:rkz047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ronneberger O, Fischer P, Brox T. U‐Net: convolutional networks for biomedical image segmentation. In: Medical image computing and computer‐assisted intervention–MICCAI 2015. Springer International Publishing: New York; 2015. p. 234–41. [Google Scholar]
- 50. Viola P, Jones M. Rapid object detection using a boosted cascade of simple features. In: Proceedings of the 2001 IEEE Computer Society Conference on Computer Vision and Pattern Recognition CVPR 2001. 2001. p. I. [Google Scholar]
- 51. Chaturvedi N. DeepRA: predicting joint damage from radiographs using CNN with attention. arXiv [cs.CV] 2021. URL: http://arxiv.org/abs/2102.06982.
- 52. Lin TY, Goyal P, Girshick R, et al. Focal loss for dense object detection. arXiv [cs.CV] 2017. URL: http://arxiv.org/abs/1708.02002. [DOI] [PubMed]
- 53. Tan M, Le QV. EfficientNet: rethinking model scaling for convolutional neural networks. arXiv [cs.LG] 2019. URL: http://arxiv.org/abs/1905.11946.
- 54. Ghassemi M, Oakden‐Rayner L, Beam AL. The false hope of current approaches to explainable artificial intelligence in health care. Lancet Digit Health 2021;3:e745–50. [DOI] [PubMed] [Google Scholar]
- 55. Lipton ZC. The mythos of model interpretability. arXiv [cs.LG] 2016. URL: http://arxiv.org/abs/1606.03490.
- 56. Sakellariou G, Conaghan PG, Zhang W, et al. EULAR recommendations for the use of imaging in the clinical management of peripheral joint osteoarthritis. Ann Rheum Dis 2017;76:1484–94. [DOI] [PubMed] [Google Scholar]
- 57. Wang X, Oo WM, Linklater JM. What is the role of imaging in the clinical diagnosis of osteoarthritis and disease management? Rheumatology (Oxford) 2018;57 Suppl:iv51–60. [DOI] [PubMed] [Google Scholar]
- 58. Tiulpin A, Klein S, Bierma‐Zeinstra SM, et al. Multimodal machine learning‐based knee osteoarthritis progression prediction from plain radiographs and clinical data. Sci Rep 2019;9:20038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Tiulpin A, Saarakkala S. Automatic grading of individual knee osteoarthritis features in plain radiographs using deep convolutional neural networks. arXiv [eess.IV] 2019. URL: http://arxiv.org/abs/1907.08020. [DOI] [PMC free article] [PubMed]
- 60. Tiulpin A, Thevenot J, Rahtu E, et al. Automatic knee osteoarthritis diagnosis from plain radiographs: a deep learning‐based approach. Sci Rep 2018;8:1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Tiulpin A, Klein S, Bierma‐Zeinstra S, et al. Deep learning predicts knee osteoarthritis progression from plain radiographs. Osteoarthritis Cartilage 2019;27:S397–8. [Google Scholar]
- 62. Kellgren JH, Lawrence JS. Radiological assessment of osteo‐arthrosis. Ann Rheum Dis 1957;16:494–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Baldi P, Chauvin Y. Neural networks for fingerprint recognition. Neural Comput 1993;5:402–18. [Google Scholar]
- 64. Altman RD, Gold GE. Atlas of individual radiographic features in osteoarthritis, revised. Osteoarthritis Cartilage 2007;15 Suppl:A1–56. [DOI] [PubMed] [Google Scholar]
- 65. Finan PH, Buenaver LF, Bounds SC, et al. Discordance between pain and radiographic severity in knee osteoarthritis: findings from quantitative sensory testing of central sensitization. Arthritis Rheum 2013;65:363–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Dejaco C, Ramiro S, Duftner C, et al. EULAR recommendations for the use of imaging in large vessel vasculitis in clinical practice. Ann Rheum Dis 2018;77:636–43. [DOI] [PubMed] [Google Scholar]
- 67. Luqmani R, Lee E, Singh S, et al. The role of ultrasound compared to biopsy of temporal arteries in the diagnosis and treatment of giant cell arteritis (TABUL): a diagnostic accuracy and cost‐effectiveness study. Health Technol Assess 2016;20:1–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Roncato C, Perez L, Brochet‐Guégan A, et al. Colour Doppler ultrasound of temporal arteries for the diagnosis of giant cell arteritis: a multicentre deep learning study. Clin Exp Rheumatol 2020;38:120–5. [PubMed] [Google Scholar]
- 69. Schmidt WA, Kraft HE, Völker L, et al. Colour Doppler sonography to diagnose temporal arteritis. Lancet 1995;345:866. [DOI] [PubMed] [Google Scholar]
- 70. Zhai X, Kolesnikov A, Houlsby N, et al. Scaling vision transformers. arXiv [cs.CV] 2021. URL: http://arxiv.org/abs/2106.04560.
- 71. Zbontar J, Jing L, Misra I. Barlow twins: self‐supervised learning via redundancy reduction. arXiv [csCV] 2021. URL: https://arxiv.org/abs/2103.03230.
- 72. Yazdany J, Bansback N, Clowse M, et al. Rheumatology informatics system for effectiveness: a national informatics‐enabled registry for quality improvement. Arthritis Care Res (Hoboken) 2016;68:1866–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Yang Q, Liu Y, Chen T, et al. Federated machine learning: concept and applications. ACM Trans Intell Syst Technol 2019;10:1–19. [Google Scholar]
- 74. Dayan I, Roth HR, Zhong A, et al. Federated learning for predicting clinical outcomes in patients with COVID‐19. Nat Med 2021;27:1735–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. McMahan B, Moore E, Ramage D, et al. Communication‐efficient learning of deep networks from decentralized data. In: Singh A, Zhu J, editors. PMLR 2017;54:1273–82. [Google Scholar]
- 76. Rieke N, Hancox J, Li W, et al. The future of digital health with federated learning. NPJ Digit Med 2020. 14;3:119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Abadi M, Chu A, Goodfellow I, et al. Deep learning with differential privacy. arXiv [stat.ML] 2016. URL: http://arxiv.org/abs/1607.00133. [Google Scholar]
- 78. U.S. Food and Drug Administration Office of the Commissioner . FDA authorizes marketing of first cardiac ultrasound software that uses artificial intelligence to guide user; 2020. URL: https://www.fda.gov/news‐events/press‐announcements/fda‐authorizes‐marketing‐first‐cardiac‐ultrasound‐software‐uses‐artificial‐intelligence‐guide‐user.
- 79. Liu X, Rivera SC, Moher D, et al, on behalf of the SPIRIT‐AI and CONSORT‐AI Working Group. Reporting guidelines for clinical trial reports for interventions involving artificial intelligence: the CONSORT‐AI extension. Nat Med 2020;26:1364–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Mitchell M, Wu S, Zaldivar A, et al. Model cards for model reporting. arXiv [cs.LG] 2018. URL: http://arxiv.org/abs/1810.03993.
- 81. Chen IY, Pierson E, Rose S, et al. Ethical machine learning in health care. arXiv [cs.CY] 2020. URL: http://arxiv.org/abs/2009.10576.
- 82. Gianfrancesco MA, Tamang S, Yazdany J, et al. Potential biases in machine learning algorithms using electronic health record data. JAMA Intern Med 2018;178:1544–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Capurro D, Coghlan S, Pires DE. Preventing digital overdiagnosis. JAMA 2022;327:525–6. [DOI] [PubMed] [Google Scholar]
- 84. Smuck M, Odonkor CA, Wilt JK, et al. The emerging clinical role of wearables: factors for successful implementation in healthcare. NPJ Digit Med 2021;4:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Holzinger A, Biemann C, Pattichis CS, et al. What do we need to build explainable AI systems for the medical domain? arXiv [cs.AI] 2017. URL: http://arxiv.org/abs/1712.09923.
- 86. Slack D, Hilgard S, Jia E, et al. Fooling LIME and SHAP: adversarial attacks on post hoc explanation methods. In: Proceedings of the AAAI/ACM Conference on AI, Ethics, and Society. New York: Association for Computing Machinery; 2020. p. 180–6. [Google Scholar]
- 87. Simonyan K, Vedaldi A, Zisserman A. Deep inside convolutional networks: visualising image classification models and saliency maps. arXiv [cs.CV] 2013. URL: http://arxiv.org/abs/1312.6034.
- 88. Adebayo J, Gilmer J, Muelly M, et al. Sanity checks for saliency maps. arXiv [cs.CV] 2018. URL: http://arxiv.org/abs/1810.03292.
- 89. Oakden‐Rayner L, Dunnmon J, Carneiro G, et al. Hidden stratification causes clinically meaningful failures in machine learning for medical imaging. Proc ACM Conf Health Inference Learn 2020;2020:151–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Caswell I, Liang B. Recent advances in google translate. Google AI Blog. URL: https://ai.googleblog.com/2020/06/recent-advances-in-google-translate.html. [Google Scholar]
- 91. Hu X, Cirit O, Binaykiya T, et al. DeepETA: how uber predicts arrival times using deep learning. Uber Engineering. URL: https://eng.uber.com/deepeta-how-uber-predicts-arrival-times/. [Google Scholar]
- 92. Chen M, Tworek J, Jun H, et al. Evaluating large language models trained on code. arXiv [cs.LG] 2021. URL: http://arxiv.org/abs/2107.03374.
- 93. Slominski A, Muthusamy V, Ishakian V. Future of computing is boring (and that is exciting!) or how to get to computing nirvana in 20 years or less. arXiv [cs.CY] 2019. URL: http://arxiv.org/abs/1906.10398.
- 94. Vig J. A multiscale visualization of attention in the transformer model. arXiv [csHC] 2019. URL: https://arxiv.org/abs/1906.05714.
- 95. Nair V, Hinton GE. Rectified linear units improve restricted Boltzmann machines. In: Proceedings of the 27th International Conference on International Conference on Machine Learning. Omnipress: Madison (Wisconsin); 2010. p. 807–14. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Disclosure Form


