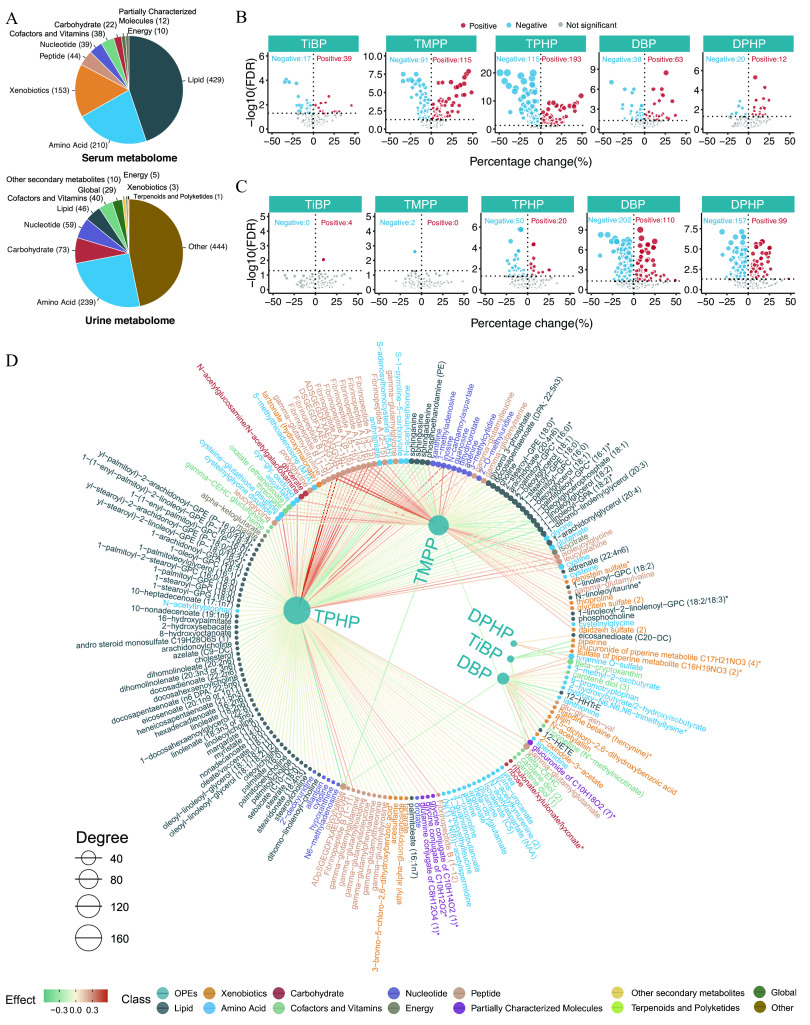
Figure 4.
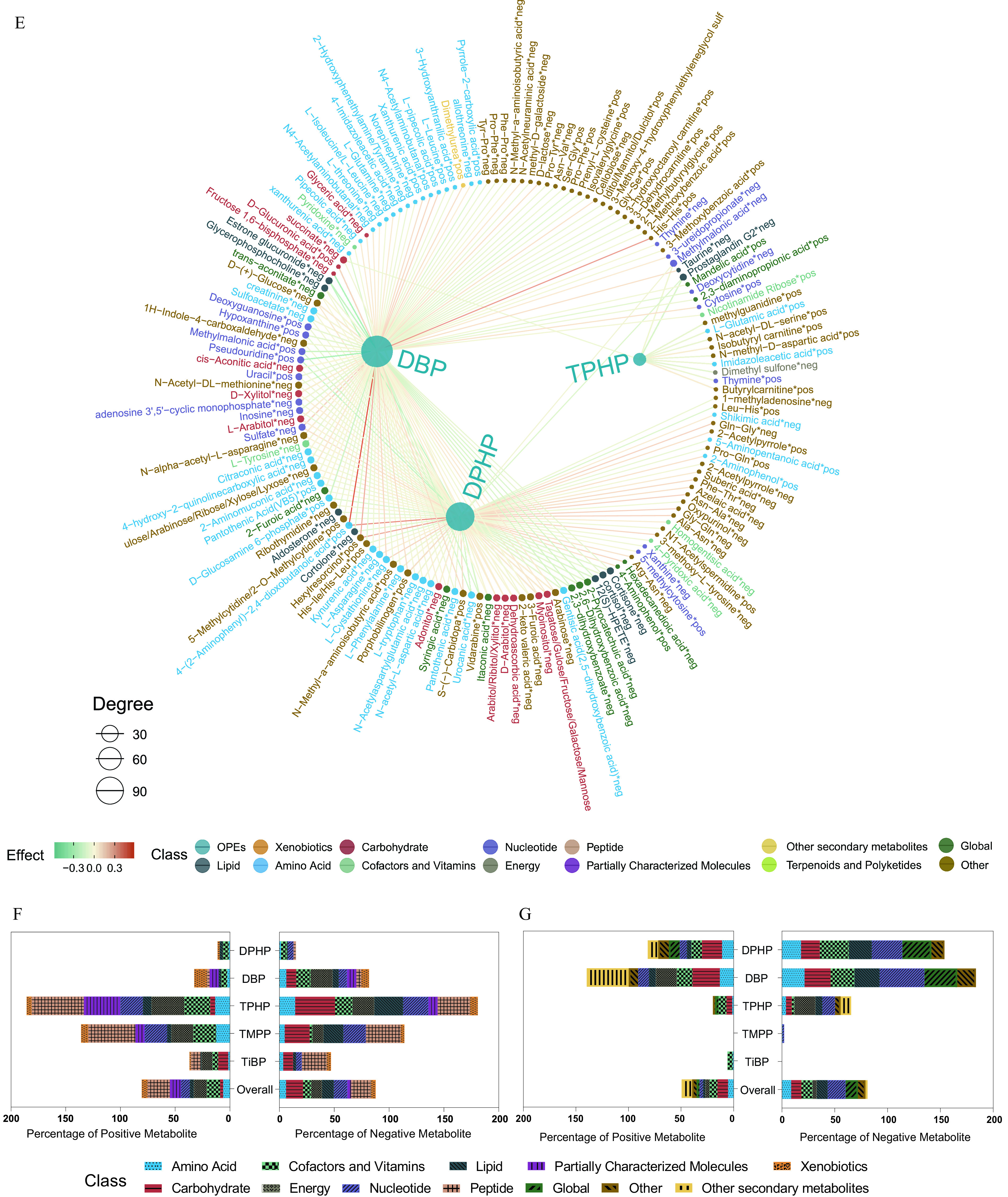
Serum and urine metabolome profiling of exposure to the key OPEs among healthy older Chinese adults 60–69 years of age. (A) The number of endogenous metabolites based on metabolic classifications in the serum and urine. (B,C) Volcano plots of the coefficient estimates of the key OPEs vs. the FDR values in the associations of the exposure–serum metabolome (B; see also Excel Table S6) and exposure–urine metabolome (C; see also Excel Table S7). Coefficient estimates are expressed as percentage changes (%) in FPG, GSP, FINS, and HOMA-IR per 1-SD change in each exposure, which was previously transformed to approach normality. The dashed horizontal line shows where the FDR value equals 0.05. (D,E) Network diagram of the association analysis between exposure to the key OPEs and the serum/urine metabolome (only metabolites with associations of FDR are shown; see also Excel Tables S6 and S7). The size of the node represents the degree of the exposure–metabolite connection, and the color of the edge represents the coefficient estimate of the exposure–metabolite association. (F,G) Stacking histograms of the percentages (%) of positively and negatively associated metabolites within each class, as well as the overall average for the serum and urine metabolomes with an FDR value of (see also Tables S5 and S6). Note: FDR, false discovery rate; FINS, fasting insulin; FPG, fasting plasma glucose; GSP, glycated serum protein; HOMA-IR, homeostatic model assessment for insulin resistance; OPE, organophosphate ester; SD, standard deviation.