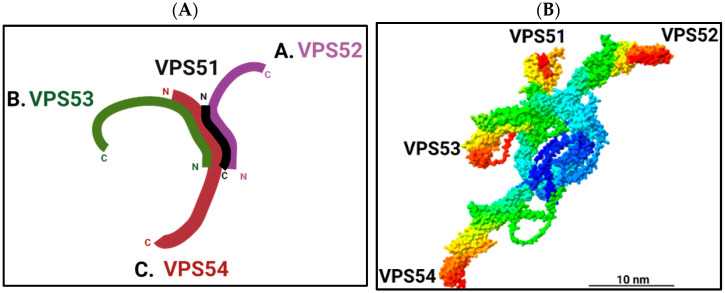
Figure 1.
Structure of the GARP complex. (A) Schematic representation of the proposed 3D structure of yeast GARP complex based on negative-stain electron microscopy [11]. (B) AlphaFold structure prediction of human GARP complex. The N-terminal regions of VPS51 (1–194), VPS52 (1–178), VPS53 (1–169), and VPS54 (1–310) were used to assemble a 3D core structure of the GARP complex using the ColabFold [20] plugin of the UCSF ChimeraX program [21]. AlphaFold predicted 3D structures of individual subunits were fitted to the core structure using the “matchmaker” command of the UCSF ChimeraX program, and the resulting structures were colored using the Rainbow palette. Scale bar is 10 nm. Predicted 3D structures of individual VPS51-54 human proteins were updated in AlphaFold DB version 2022-11-01, created with the AlphaFold Monomer v2.0 pipeline. The predicted 3D structure of the GARP complex was assembled on 27 December 2022 and colored using ChimeraX.