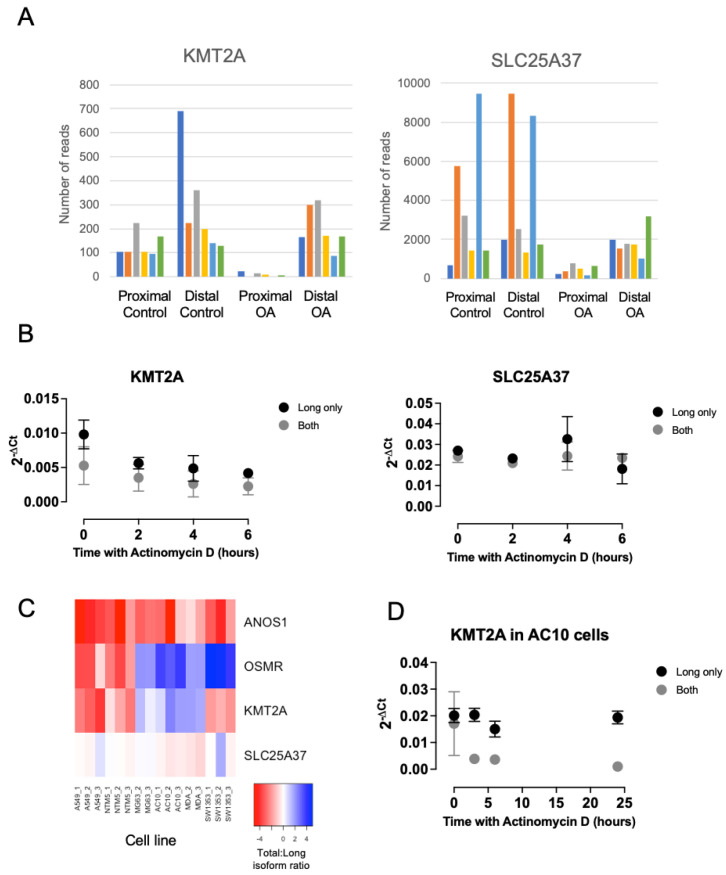
Figure 5.
(A) QuantSeq-Reverse read numbers for proximal and distal polyadenylation sites for the KMT2A and SLC25A37 genes, usage of which leads to different length 3′UTRs and is differentially regulated between control and osteoarthritic (OA) samples. KMT2A proximal site is at CHR11:118523915, KMT2A distal site is at CHR11:118526830, SLC25A37 proximal site is at CHR8:23,572,547, SLC25A37 distal site is at CHR8:235,75,460. Different colour bars represent read numbers from individual control or OA samples allowing per sample comparison between proximal and distal values in each case. (B) RT-qPCR analysis of mRNA decay rates of KMT2A and SLC25A37 using primer pairs that are specific to only longer 3′UTR isoforms (long only) or both proximal and ideally polyadenylated isoforms (both) in human articular chondrocytes treated with actinomycin D. Mean ± standard error shown, n = 3. (C) Heat map showing ratio of expression of intronic to canonical 3′UTR (for OSMR and ANOS1) or long only to both for (KMT2A and SLC25A37) in a panel of different cell lines. (D) RT-qPCR analysis of mRNA decay rates of long only or both polyadenylated isoforms of KMT2A in AC10 cells. Mean ± standard error shown, n = 3.