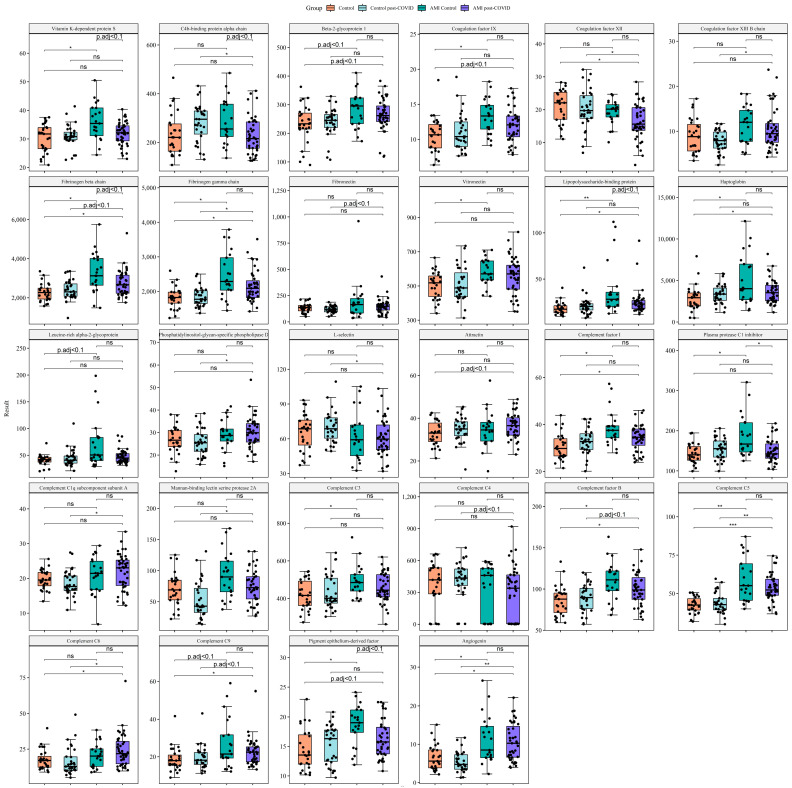
Figure 1.
Main comparisons of proteomics data between study groups. Targeted proteomic analysis was carried out using liquid chromatography–tandem mass spectrometry (LC-MS/MS) with multiple-reaction monitoring (MRM). For comparison of several groups, we used the Mann–Whitney rank test with a continuity correction. In order to overcome errors from multiple comparisons, we performed a Benjamini–Hochberg FDR correction, with calculation of critical values for each comparison matched with corresponding p-values; we calculated adjusted p-values and compared them with a critical value of 0.1 to keep the positive false discovery rate below 10%. Data are presented as the median [Q1; Q3]. * p adj. < 0.05, ** p adj. < 0.01, *** p adj. < 0.001, ns, not significant. The boxplots include proteins different between the AMI control and AMI post-COVID groups: C4b-binding protein alpha-chain, fibrinogen beta-chain, lipopolysaccharide-binding protein, PEDF, plasma protease C1 inhibitor, vitamin-K-dependent protein S. Based on their function, we focused on proteins connected to hemostasis, acute-phase proteins, and components of the complement system. In the boxplots, we included proteins with these functions if they were different in at least one of the comparisons: AMI control vs. control, AMI post-COVID vs. control, AMI post-COVID vs. control post-COVID.