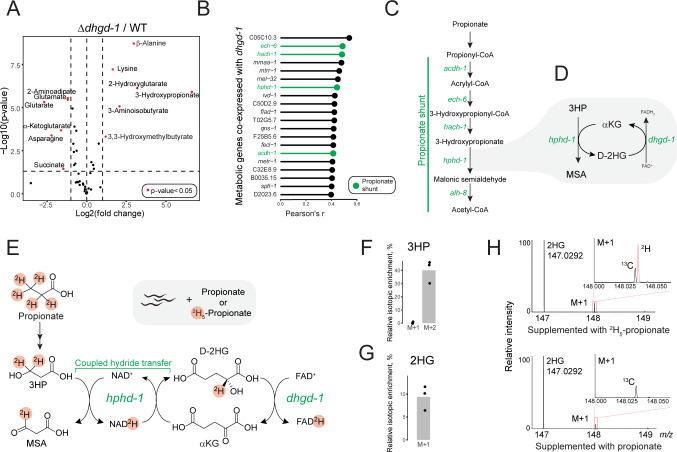
Fig 2. Dhgd-1 functions in the propionate shunt pathway.
(A) GC-MS profiling of metabolic changes in Δdhgd-1 mutants compared to WT C. elegans. P-values are Benjamini–Hochberg adjusted. (B) Metabolic genes most highly coexpressed with dhgd-1. Propionate shunt pathway genes are enriched with an FDR of 0.042 (S2 Table). (C, D) Proposed mechanism of D-2HG production (by HPHD-1) and recycling (by DHGD-1) during propionate degradation via the propionate shunt pathway. (E) Isotope tracing experiment with deuterium (2H)-labeled propionate and proposed reaction mechanism. HPHD-1 uses NAD+/NADH as a hydride shuttle in a coupled reaction yielding 1 equivalent each of MSA and D-2HG. (F, G) Fractional enrichment of 3HP (F) and 2HG (G) isotopologues. Bars indicate mean of n = 3 biological replicates. (H) HPLC-MS analysis of the M+1 isotope cluster in animals fed 2H5-propionate revealed robust incorporation of a single deuterium atom in 2HG, whereas the M+1 in animals fed propionate was exclusively from natural abundance of 13C. The data underlying Fig 2F and 2G can be found in S1 Data. D-2HG, D-2-hydroxyglutarate; FDR, false discovery rate; GC-MS, gas chromatography–mass spectrometry; HPLC-MS, high performance liquid chromatography mass spectrometry; MSA, malonic semialdehyde; WT, wild type.