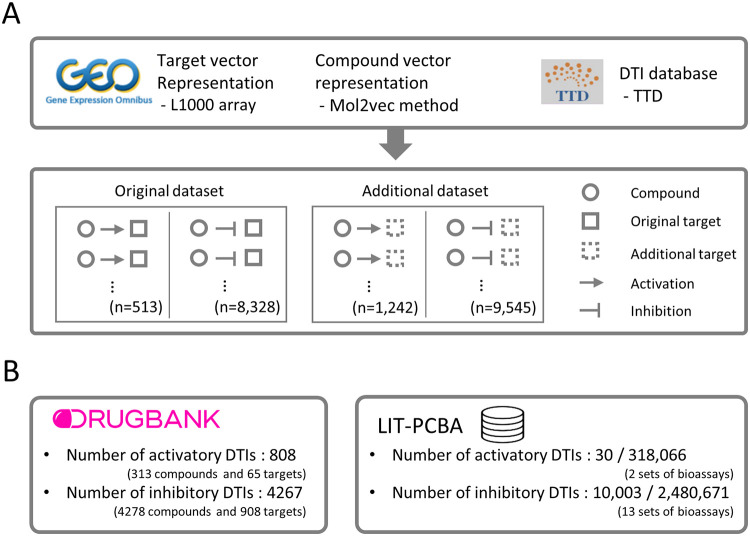
Fig 3. Construction of a dataset of known DTIs and features.
(A) Training dataset construction. Transcriptome profiles were obtained from the L1000 array data and then aggregated to generate a representative target vector. A mol2vec method was used to generate representative vectors for compounds. DTIs with modes of action were collected from the TTD. The original dataset was constructed by selecting activatory and inhibitory DTI pairs that include a compound for which ECFPs can be calculated and an original target (i.e., a target for which genetically perturbed transcriptome data are available). The additional dataset was constructed by selecting activatory and inhibitory DTI pairs that include a compound for which ECFPs can be calculated and an additional target (i.e., a target for which inferred transcriptome data are available). (B) Independent dataset construction. Two independent datasets, Drugbank and LIT-PCBA datasets, were constructed to evaluate the reliability of predictions for unseen DTI in training datasets.