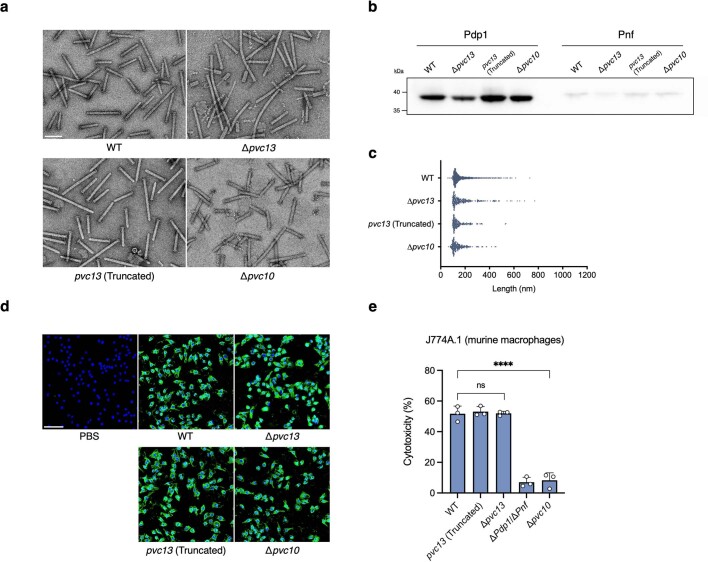
Extended Data Fig. 7. Properties of PVCs containing mutant tail fibre (Pvc13) or spike tip (Pvc10).
a, PVCs harbouring deleted pvc13 (as was used in Fig. 1f, g), truncated pvc13 (lacking aa 403-476, the binding domain characterized in Fig. 2a), or deleted pvc10 (as was used in Fig. 4b–e) all still form intact particles under TEM, indicating these genes are not necessary for the assembly of the PVC complex. Scale bar, 100 nm. b, Modifications to pvc13 or pvc10 do not affect the loading of payload proteins. Endogenous PVC payloads Pdp1 and Pnf were tagged with HiBiT and loaded into either wild-type or mutant PVC complexes; successful payload loading was assessed with a HiBiT blot. No significant depletion of Pdp1 or Pnf was observed in the mutant PVCs, indicating these genes are not necessary for payload loading. c, PVC length regulation is not disrupted by modifications to pvc13 or pvc10. PVC lengths were measured via a similar technique as in Extended Data Fig. 1b; no significant change in length distribution was observed in the mutant PVCs. d, PVCs bind to mouse macrophages in the absence of wild-type pvc13 or pvc10. J774A.1 cells (a cell line used in previous studies of PVC activity1–3) were exposed to PVCs tagged with Flag (as in Fig. 1d) and binding was assessed with immunofluorescence. PVCs harbouring modified Pvc13 still clustered on the surface of J774A.1 cells, indicating this binding interaction was not mediated by Pvc13 in the same way that it was for human cells in Fig. 2a. Scale bar, 100 μm. e, PVC activity in mouse macrophages persists in the absence of wild-type pvc13. In support of the results from (d), PVCs containing truncated or deleted pvc13 produced efficient cytotoxicity in J774A.1 cells, indicating PVC activity in this cell line is not the result of specific recognition by the tail fibre. However, deleting the spike tip (pvc10) does produce a loss of activity, suggesting the PVC may nonetheless be nonspecifically active against these cells. Values are mean ± s.d. with n = 3 biological replicates. Statistical significance was computed using one-way ANOVA with Bonferroni post hoc test; ****P < 0.0001; ns, not significant.