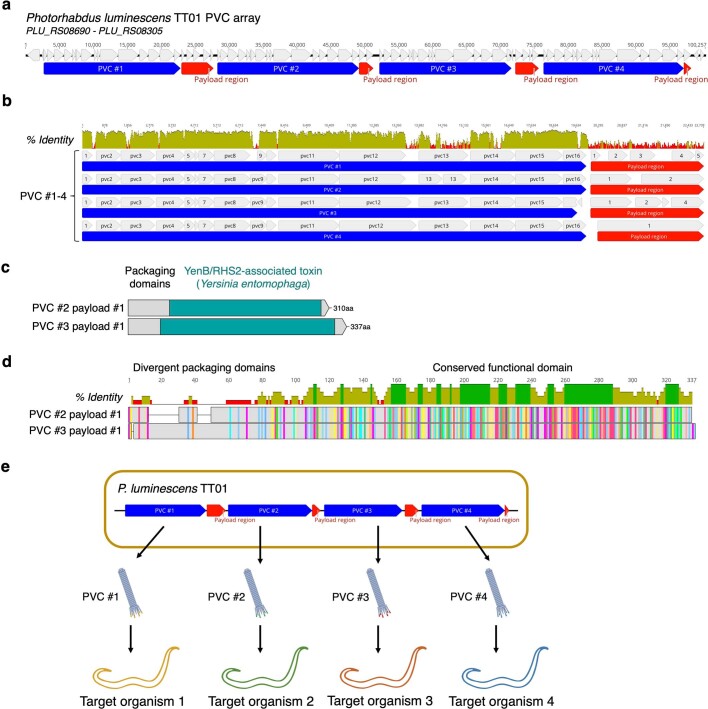
Extended Data Fig. 9. The PVC array in P. luminescens TT01 reveals a multiplexed protein delivery strategy.
a, P. luminescens TT01 contains an array of four PVC loci in tandem on the genome, with each structural/accessory region (blue) immediately followed by putative payload genes (red). b, DNA sequence alignment of the four PVC loci in the P. luminescens TT01 PVC array. The loci share significant sequence homology but diverge at the target recognition gene (pvc13) and the payload region. This suggests these PVCs share a core structure but target different organisms and deliver different payloads to each target. Percent identity is presented with a sliding window of n = 50 bp. c, HHpred-predicted domain organization of two putative payload genes shared by separate PVC loci in the P. luminescens TT01 PVC array. While both genes share a common domain (YenB/RHS2-associated toxin; E = 0.0000067/0.0000039), both also contain unknown sequences on the N termini. Given that the PVCpnf payloads Pdp1 and Pnf are loaded via N-terminal packaging domains (Extended Data Fig. 3b–d), it is likely these N-terminal domains represent packaging domains which load these proteins into PVC complexes. d, Amino acid sequence alignment of putative payload proteins from (c). The two payloads share significant sequence homology in the functional domain (YenB/RHS2-associated toxin) but diverge at the N termini. It is plausible these two payloads are “sorted” into different PVC particles within P. luminescens TT01 via their divergent N-terminal packaging domains. Percent identity is presented with a sliding window of n = 5 aa. e, Proposed model for multiplexed protein delivery by P. luminescens TT01. This organism likely produces four distinct PVC particles, each with a different payload (sorted into each particle via unique packaging domains) and a different targeting apparatus (pvc13), which could be secreted to effect divergent biological activity within multiple host organisms (or cell types/tissues) simultaneously.