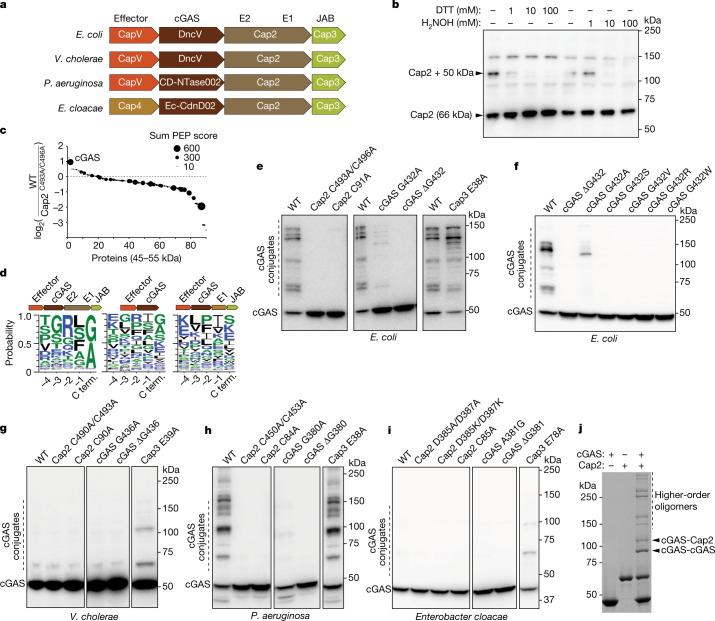
Fig. 1. Cap2 catalyses covalent conjugation of cGAS.
a, Domain organization of the CBASS operons from E. coli, Vibrio cholerae, Pseudomonas aeruginosa and Enterobacter cloacae. b, E. coli MG1655, which lacks CBASS, was transformed with the four-gene operon (from a) including an N-terminally Flag-tagged Cap2. Bacterial cell lysates were treated for 30 min with the indicated amount of DTT or hydroxylamine (H2NOH), subjected to non-reducing SDS–PAGE and immunoblotted with an anti-Flag antibody. c, Mass spectrometry analysis of 45–55 kDa proteins conjugated to Cap2 that migrated at about 110 kDa on an SDS–PAGE gel. The y axis indicates enrichment of the proteins in bacteria with wild-type CBASS compared to those harbouring the C493A/C496A mutations of Cap2. The sum PEP score is the sum of negative logarithms of the posterior error probabilities (PEP) and represents the credibility of the spectral matches. d, Sequence logo of alignment of the C-terminal (C term.) sequences of cGAS from previously identified CBASS operons12 with the indicated gene architecture: operons containing full-length Cap2 and Cap3 (left), minimal operons with no Cap2 or Cap3 (centre) and operons lacking the E2 domain in Cap2 (right). e–i, Immunoblotting of cGAS conjugates in bacterial lysates from cells harbouring the wild-tye (WT) four-gene CBASS operon with N-terminally Flag-tagged cGAS, or those with indicated mutations in the CBASS (e) and cGAS (f) genes from E. coli, V. cholerae (g), P. aeruginosa (h) and Enterobacter cloacae (i). Bacteria cell lysates were subjected to SDS–PAGE in the presence of β-mercaptoethanol, followed by immunoblotting with an anti-Flag antibody. j, Purified recombinant cGAS and Cap2 proteins were incubated in the presence of ATP for 30 min at room temperature, as indicated. Following the reaction, proteins were separated by SDS–PAGE and stained with Coomassie blue. All data are representative of at least two independent experiments. For gel source data, see Supplementary Fig. 1.