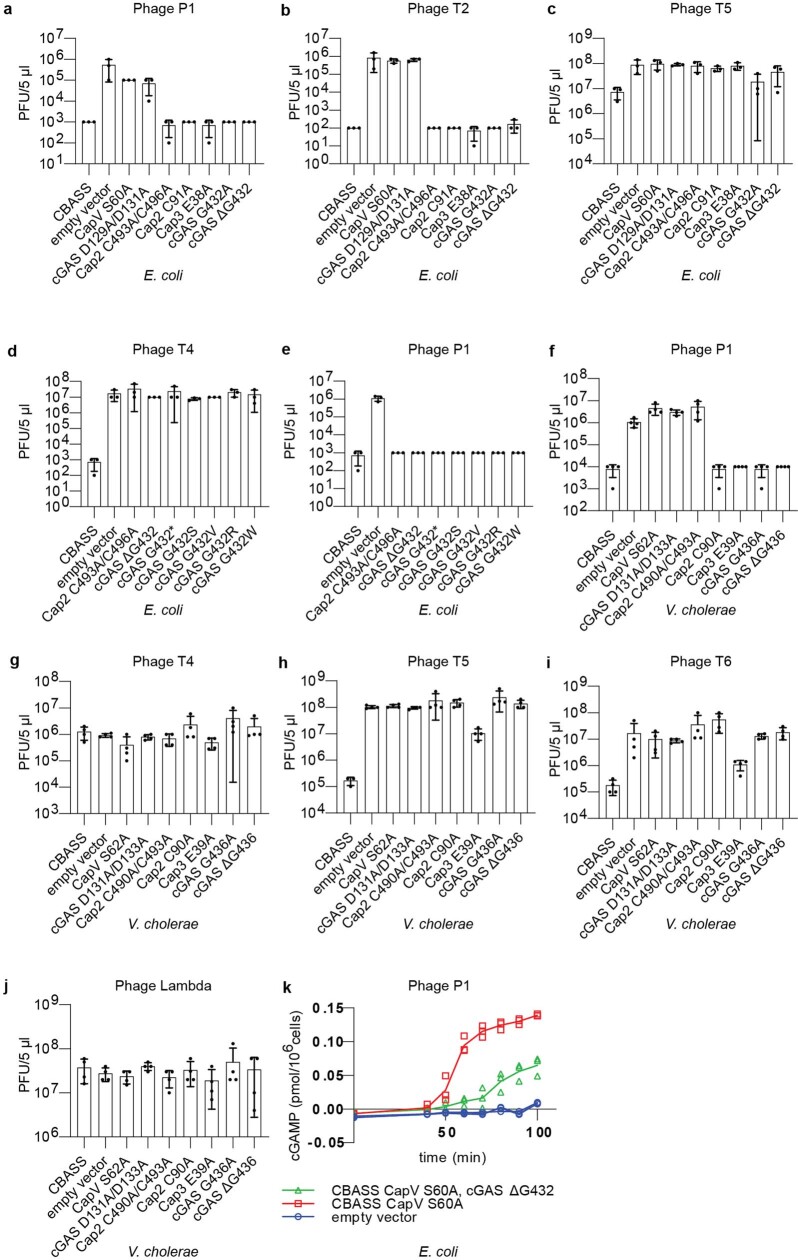
Extended Data Fig. 2. Viral titers of bacteriophages in bacteria harboring CBASS operons with point mutations in the indicated genes.
a–c, E. coli MG1655 was transformed with the wild-type operon from E. coli TW11681 (CBASS) and the operon containing the indicated mutations. Empty vector indicates no CBASS operon. The strains were infected with Phage P1 (a), Phage T2 (b), or Phage T5 (c) and the plague forming units (PFU) were measured. Bar graph represents mean ± SD of n = 3 independent experiments for each bacterial strain. d–e, Viral titers of phage T4 (d) and P1 (e) after infection in bacterial strains harboring the wild-type CBASS operon from E. coli TW11681, no operon (empty vector), or the indicated point mutations. Bar graph represents average values ± SD of n = 3 independent experiments with individual points overlaid. f–j, E. coli MG1655 was transformed with the wild-type operon from V. cholerae (CBASS) with Flag-tagged cGAS and the indicated mutations. Empty vector indicates no CBASS operon. The strains were infected with Phage P1 (f), Phage T4 (g), Phage T5 (h), Phage T6 (i), or Phage Lambda (j) and the plague forming units (PFU) were measured. Bar graph represents mean ± SD of n = 4 independent experiments for each bacterial strain. k, E. coli harboring the indicated CBASS operon was infected by the phage P1 at a multiplicity of infection of 2. CapV in the CBASS operon was inactivated by a mutation in the active site (S60A) to inhibit cell death from CBASS signaling. Samples from each bacterial culture were collected at 10-min intervals starting at 40 min after infection, snap frozen, and lysed by heating and sonication. Clarified lysates were incubated with purified CapV in the presence of resorufin butyrate, a fluorogenic phospholipase substrate, to measure the bioactivity of cGAMP produced by the bacteria. Scatter plot represents mean ± SD of n = 3 technical replicates with individual points overlaid and is representative of two independent experiments.