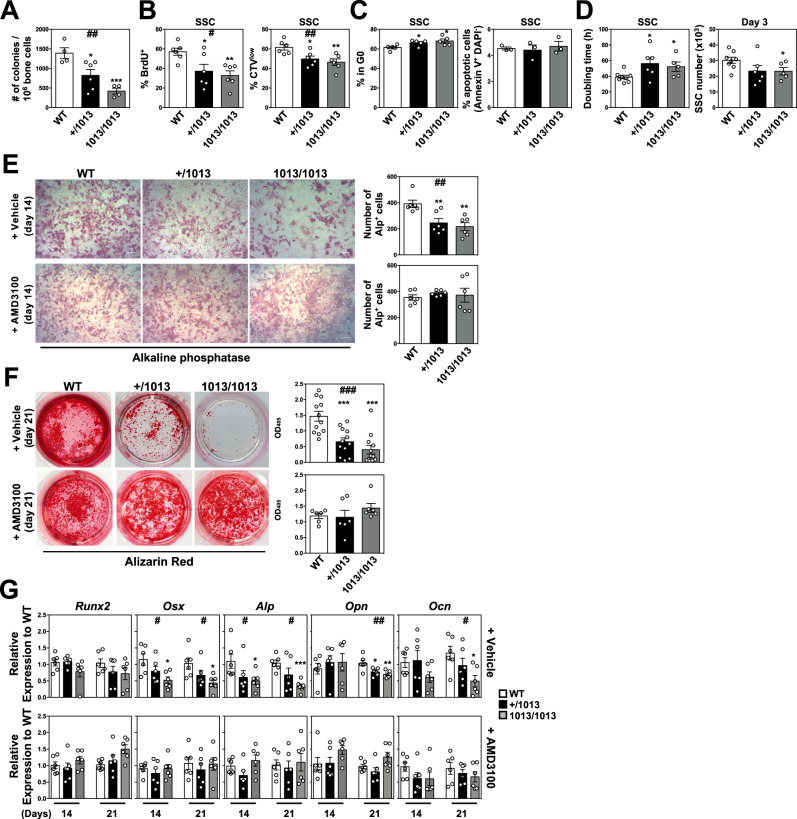
Fig. 6. Cxcr4 desensitization intrinsically regulates in vitro the osteogenic differentiation of skeletal stromal cells.
A Number of colonies formed from bone fractions in CFU-F assays. Data (means ± SEM) are from two independent experiments with n = 4, 6, and 4 mice in total for WT, +/1013, and 1013/1013 groups, respectively. Statistics were calculated with the nonparametric Kruskal–Wallis H test (##p = 0.002) and the unpaired two-tailed Student’s t test (+/1013 vs WT *p = 0.029, 1013/1013 vs WT ***p = 0.0008). B After in vitro loading with BrdU (5 days) or CTV (3 days), the percentages of BrdU+ (left) or CTVlow (right) cells within WT and mutant bone-derived SSCs were determined by flow cytometry. Data (means ± SEM) are from 3 independent experiments with 6 mice in total per group. Statistics were calculated with the nonparametric Kruskal–Wallis H test (#p = 0.0231 and ##p = 0.0047 for BrdU+ and CTVlow, respectively) and the unpaired two-tailed Student’s t test (+/1013 vs WT *p = 0.031, 1013/1013 vs WT **p = 0.0023 for BrdU+; +/1013 vs WT *p = 0.015, 1013/1013 vs WT **p = 0.0054 for CTVlow). C Bar graphs show the percentages of cultured WT or mutant SSCs in the quiescent G0 phase (DAPIlowKi-67−, left) or with an apoptotic phenotype (Annexin V+ DAPI-, right) as determined by flow cytometry. Data (means ± SEM) are from three (right panel) or five (left panel) independent SSC cultures per genotype. Statistics were calculated using the unpaired two-tailed Student’s t test (+/1013 vs WT *p = 0.032, 1013/1013 vs WT *p = 0.021). D Doubling time (left) and absolute numbers (right) of WT and mutant SSCs after 3 days of culture. Data (means ± SEM) are from 8, 6, and 5 independent SSC cultures for WT, +/1013, and 1013/1013 groups, respectively. Statistics were calculated using the unpaired two-tailed Student’s t test (+/1013 vs WT *p = 0.028, 1013/1013 vs WT *p = 0.033 for doubling-time; 1013/1013 vs WT *p = 0.048 for SSC). E Alkaline phosphatase (Alp) staining was performed 14 days after initiation of the culture of WT and mutant SSCs in osteogenic medium supplemented every two days with 10 μM AMD3100 or vehicle (PBS) (bars: 100 μm). Quantitative analyses (number of Alp+ cells) were performed under an inverted microscope. Data (means ± SEM) are from 6 independent cultures per genotype. Statistics were calculated with the nonparametric Kruskal–Wallis H test (##p = 0.0022) and the unpaired two-tailed Student’s t test (+/1013 vs WT **p = 0.0075, 1013/1013 vs WT **p = 0.0018). F Alizarin Red staining was performed 21 days after initiation of the culture. Quantitative analyses (means ± SEM) of staining were performed using the osteogenesis assay kit in 12 (vehicle) or 6 (AMD3100) independent cultures per genotype. Statistics were calculated with the nonparametric Kruskal–Wallis H test (###p = 0.0002) and the unpaired two-tailed Student’s t test (+/1013 vs WT ***p = 0.0005, 1013/1013 vs WT ***p < 0.0001). G Expression levels of osteogenic genes were determined by quantitative PCR in 6 independent WT and mutant SSC cultures 14 and 21 days after initiation of the osteogenic culture in the presence or absence of AMD3100. Each individual sample was run in triplicate and was standardized for β-actin expression levels. Results (means ± SEM) are expressed as relative expression compared to WT samples. Statistics were calculated with the nonparametric Kruskal–Wallis H test (#p = 0.038 and 0.0205 for Osx days 14 and 21, respectively; #p = 0.024 and 0.015 for Alp days 14 and 21, respectively; ##p = 0.0063 for Opn days 21; #p = 0.026 for Ocn days 21) and the unpaired two-tailed Student’s t test (1013/1013 vs WT *p = 0.0107 and 0.013 for Osx days 14 and 21, respectively; 1013/1013 vs WT *p = 0.035 and ***p = 0.0001 for Alp days 14 and 21, respectively; +/1013 vs WT *p = 0.022 and 1013/1013 vs WT **p = 0.0056 for Opn days 21; 1013/1013 vs WT **p = 0.0108 for Ocn days 21). All mice were littermates, females, and age-matched (8–12 wk-old). Source data are provided as a Source data file.