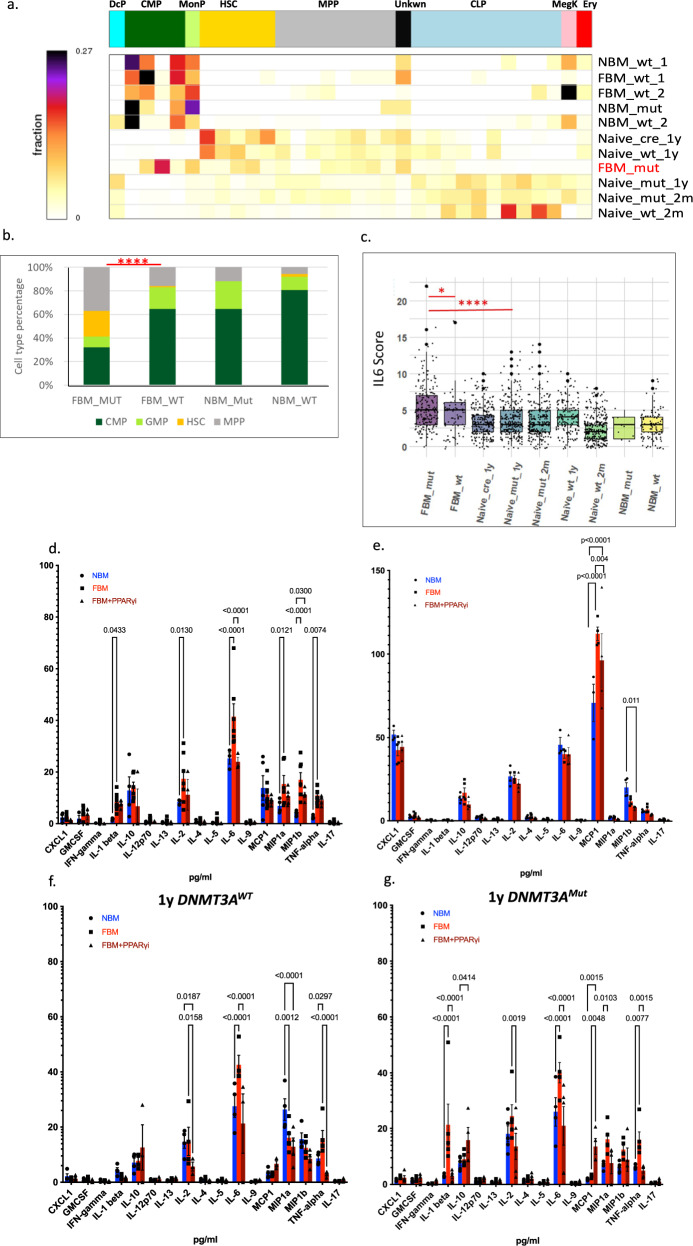
Fig. 4. DNMT3AMut cells exposed to FBM maintain an HSC pool characterized by an inflammatory phenotype.
a Cells from DNMT3AMut and DNMT3AWT were injected to mice (n = 3) with fatty bone marrow (FBM) and normal bone marrow (NBM). Three days after injection lin- KIT+ (LK) cells were isolated from mice bone marrow (BM) and underwent single-cell RNA-Seq analysis. MetaCell algorithm was used to assign different single cells to Metacells with unique gene programs and cell types44. Gold, hematopoietic stem cells (HSCs); dark green, common myeloid progenitors (CMP); light blue, common lymphoid progenitors, (CLP); cyan, dendritic progenitors (DcP); gray, multipotent progenitors (MPP); dark olivegreen, monocyte progenitors (MonoP); pink, megakaryocyte progenitors (MegK); red, erythroid progenitors (Ery); grey4 (Unknown). Conditions: normal bone marrow (NBM); wild-type (wt); fatty bone marrow (FBM); naive cells—are cells extracted directly from BM of respective mice without transplantation. cre is the cre control. b While all injected cells show a marked reduction in HSCs after transplantation DNMT3AMut cells exposed to FBM maintain their HSC pool which is significantly higher than all other condition (which were transplanted). Exact fisher test was used ****P < 0.0005 to compare proportions of HSCs between the groups. FBM_MUT vs. FBM_WT P = 2.3 e10−9. c Ranked GSEA analysis on differentially expressed genes between DNMT3AMut cells exposed to FBM cluster and other clusters exposed significant enrichment of inflammatory pathways one of them was the IL-6, JAK and STAT3 response gene set. An expression score for every single cell was calculated based on the expression of each of the genes in the IL-6 gene set. All comparisons were performed using a two-tailed, nonpaired, nonparametric Wilcoxon rank-sum test with 95% confidence interval with FDR multiple hypothesis. *P < 0.05, ****P < 0.0005. FBM_mut vs. FBM_wt P = 0.01281, FBM_mut vs. Naive_mut_1y P = 2.18E-09. d Multiplex cytokines assay (FirePlex-96 Key Cytokines (Mouse) Immunoassay Panel (ab235656)) of 17 common cytokines analyzed by FACS-based multiplex method of serum from NBM, FBM and following PPARγi administration to NSG mice, without any cell’s transplantation. Data were analyzed by two-way ANOVA test—Sidaks multiple comparison test. The figure displays the P values. NBM n = 4, 3, 3, 4, 4, 4, 4, 3, 4, 4, 3, 4, 6, 4, 5, 4, 6. FBM n = 9, 7, 6, 9, 6, 6, 7, 7, 8, 8, 7, 9, 9, 6, 7, 9, 8. FBM+PPARγi n = 5, 3, 3, 5, 3, 3, 3, 3, 3, 3, 3, 3, 5, 5, 5, 5, 3. e Multiplex cytokines assay (FirePlex-96 Key Cytokines (Mouse) Immunoassay Panel (ab235656)) of 17 common cytokines analyzed by FACS-based multiplex method of BM from NBM, FBM and following PPARγi administration to NSG mice, without any cell’s transplantation. Data were analyzed by two-way ANOVA test—Sidaks multiple comparison test. The figure displays the P values. NBM n = 3, 4, 4, 4, 4, 4, 4, 4, 4, 4, 3, 4, 3, 4, 4, 4, 4. FBM n = 5, 5, 5, 5, 5, 5, 5, 3, 5, 5, 3, 4, 4, 4, 5, 5, 4. FBM+PPARγi n = 4 for all. f, g FACS-based multiplex method of NBM, FBM, and following PPARγi administration to NSG mice transplanted with 1-year-old DNMT3AMut or DNMT3AWT BM-derived cells. Each bar represents 4 to 5 mice. Data were analyzed by two-way ANOVA test—Sidaks multiple comparison test. The figure displays the P values. f NBM n = 4 for all. FBM n = 4, 4, 4, 4, 4, 4, 4, 4, 4, 5, 4, 5, 5, 5, 5, 5, 4. FBM+PPARγi n = 4, 3, 3, 4, 3, 3, 3, 3, 3, 3, 3, 3, 3, 4, 4, 4, 3. g NBM n = 5 for all, except for IL-6 n = 4. FBM n = 5 for all. FBM+PPARγi n = 5, 3, 3, 5, 4, 3, 5, 5, 5, 5, 5, 5, 4, 3, 3, 5, 5. Source data are provided as a Source Data file.