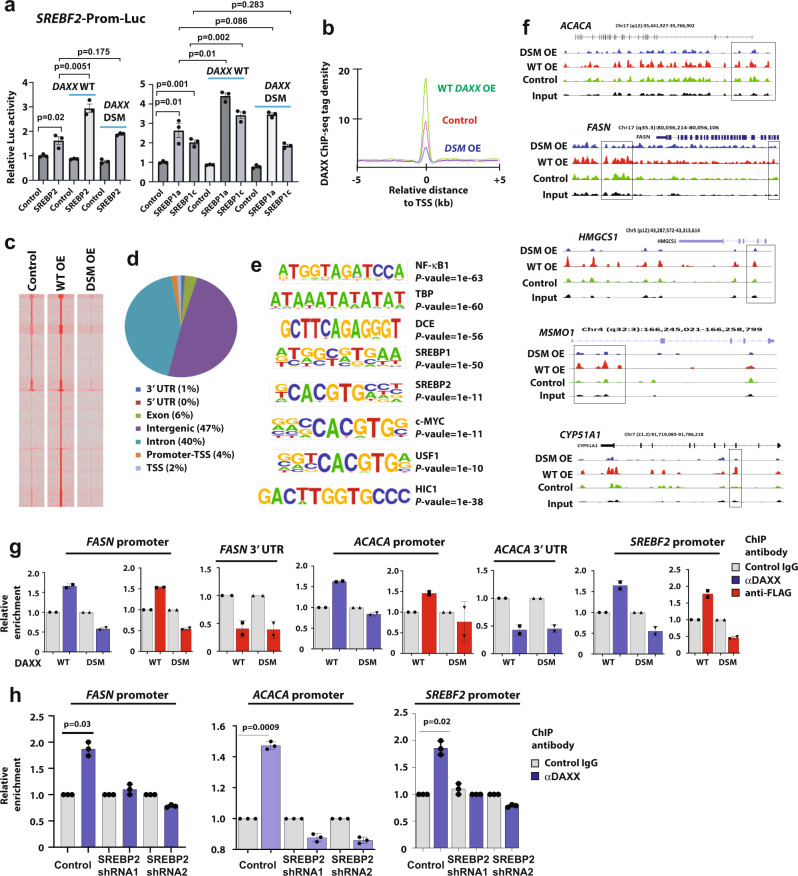
Fig. 4. DAXX activates SREBP-mediated transcription and occupies the promoters of lipogenic genes.
a MDA-MB-231 cells were transfected with a luciferase reporter driven by a promoter fragment from the SREBF2 gene along with mature SREBP2, SREBP1a, SREBP1c, wt DAXX or the DSM mutant cDNA as indicated. Dual luciferase assays were done. Data are presented as mean values ± SEM (n = 3 independent transfections). Luc: luciferase. b–f ChIP-seq analysis of genome-wide occupancy of DAXX. b, c ChIP-seq signal intensity plot (a comparison of the average DAXX ChIP-seq tag intensities) and heatmaps in MDA-MB-231 control, WT OE, and DSM OE cell lines; signals are centralized to transcriptional start sites (TSS). d The genome-wide distribution of DAXX chromatin occupancy. e Motifs enriched as determined by the DAXX ChIP-seq dataset of MDA-MB-231 WT OE cells, and (f) occupancy of WT and the DSM mutant DAXX in selected lipogenic genes based on ChIP-seq data. Boxes in panel f highlight regions near the 5’ end of each gene along with a region 3’ to the FASN gene with notable differences in peak heights between WT DAXX and the DSM mutant. In e, the HOMER software uses ZOOPS scoring (zero or one occurrence per sequence) coupled with the hypergeometric enrichment calculations (or binomial) to determine the P value for motif enrichment. g MDA-MB-231 cells stably expressing WT DAXX, and the DSM mutant were subjected to ChIP with a control IgG, an anti-DAXX (5G11), or an anti-FLAG antibody (all the DAXX constructs carrying an N-terminal FLAG epitope tag). The precipitated DNAs were subjected to qPCR with primers specific to the promoter regions of the indicated genes. qPCRs for the 3’ UTR of the FASN and ACACA genes serve as negative controls (n = 2 independent ChIP experiments). h SREBP2 is critical for DAXX to bind lipogenic gene promoters. MDA-MB-231 cells with a control vector or a SREBP2 shRNA vector were subjected to ChIP with a control IgG, or an anti-DAXX (5G11) antibody followed by qPCR with primers specific to the indicated gene promoters (n = 3 independent ChIP experiments). P values are based on unpaired two-tailed t test. Source data are provided as a Source Data file.