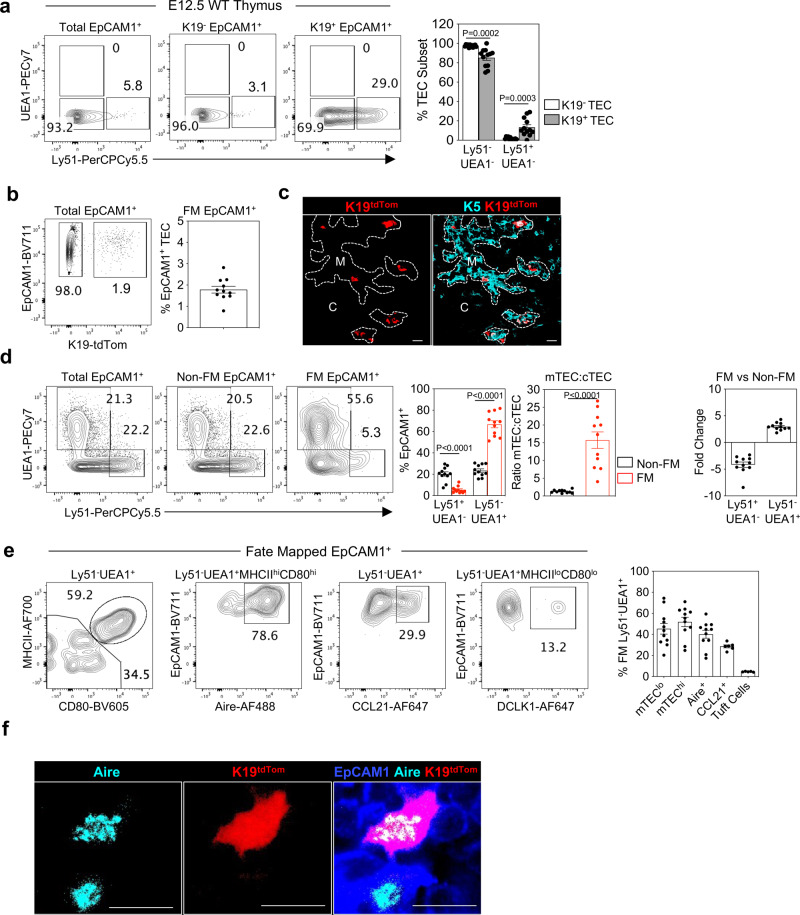
Fig. 7. K19+ mmTECp are present at E12.5 of development.
a Representative FACS plots showing expression of UEA1 and Ly51 by total EpCAM1+ cells, K19-EpCAM1+ cells and K19+EpCAM1+ cells at E12.5, and corresponding quantitation, n = 12, from 3 independent experiments. b Krt19CreERTtdTom embryos were fate-mapped via tamoxifen administration at E12.5, and the neonatal thymus was harvested at birth. Fate-mapped cells were quantitated (n = 11, from 3 independent experiments) c Immunofluorescence of fate-mapped thymi at PNd0, K5 (turquoise) K19-tdTom (red), Scale bar denotes 20um. Image representative of 4 thymi, from 3 independent experiments. d Expression of Ly51 and UEA1 by total EpCAM1+ cells, K19-Tom- EpCAM1+ cells (non-fate-mapped), and K19-tdTom+EpCAM1+ cells (fate-mapped) at PNd0, bar charts show corresponding quantitation, n = 11, from 3 independent experiments. e Representative FACS plots illustrating the phenotype of fate-mapped mTEC subsets within PNd0 fate-mapped thymus. mTEChi (MHCIIhiCD80hi, n = 11), mTEClo (MHCIIloCD80lo, n = 11), Aire+ (MHCIIhiCD80hiAire+, n = 11), CCL21+ (n = 6) and tuft cells (MHCIIloCD80loDCLK1+, n = 5) and corresponding quantitation. f Immunofluorescence of fate-mapped thymi at PNd0, Aire (turquoise) K19-tdTom (red), EpCAM1 (blue). Scale bar,10 µm. Image representative of 4 thymi. Data analysed using a Student’s t test. The data are shown as mean ± SEM.