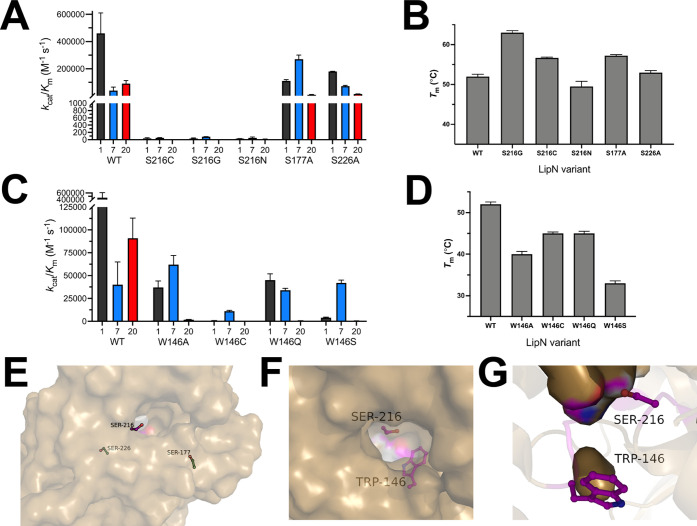
Figure 4.
Detailed analysis of two key binding pocket residues. Kinetic analysis of MmLipN variants at Ser216 (A) and Trp146 (C) against substrates 1, 7, and 20 (Figure 1A). Thermal stability of MmLipN variants at Ser216 (B) and Trp146 (D). All measurements were completed as outlined in Figure 3 and are shown ± SD. Detailed biochemical analysis of Ser216 and Trp146 is provided in Tables S4 and S5, respectively. (E) Surface structure of MmLipN illustrating the distant location of control serine substitutions (Ser177 and Ser226; green) from the nucleophilic serine (Ser216, magenta). (F, G) Location of Trp146 in relation to the nucleophilic serine (Ser216) from the exterior surface (F) and interior surface (G) of the binding pocket.