Abstract
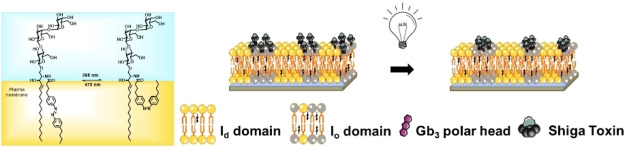
The plasma membrane is a complex assembly of proteins and lipids that can self‐assemble in submicroscopic domains commonly termed “lipid rafts”, which are implicated in membrane signaling and trafficking. Recently, photo‐sensitive lipids were introduced to study membrane domain organization, and photo‐isomerization was shown to trigger the mixing and de‐mixing of liquid‐ordered (l o) domains in artificial phase‐separated membranes. Here, we synthesized globotriaosylceramide (Gb3) glycosphingolipids that harbor an azobenzene moiety at different positions of the fatty acid to investigate light‐induced membrane domain reorganization, and that serve as specific receptors for the protein Shiga toxin (STx). Using phase‐separated supported lipid bilayers on mica surfaces doped with four different photo‐Gb3 molecules, we found by fluorescence microscopy and atomic force microscopy that liquid disordered (l d) domains were formed within l o domains upon trans‐cis photo‐isomerization. The fraction and size of these l d domains were largest for Gb3 molecules with the azobenzene group at the end of the fatty acid. We further investigated the impact of domain reorganization on the interaction of the B‐subunits of STx with the photo‐Gb3. Fluorescence and atomic force micrographs clearly demonstrated that STxB binds to the l o phase if Gb3 is in the trans‐configuration, whereas two STxB populations are formed if the photo‐Gb3 is switched to the cis‐configuration highlighting the idea of manipulating lipid‐protein interactions with a light stimulus.
Keywords: azobenzene, glycosphingolipids, membrane domains, photo-pharmacology, Shiga toxin
Switch it on: Gb3 sphingolipids with azobenzene groups serve as light‐triggered nano‐tools to control lipid and protein domains. By using the ability of Gb3 to bind Shiga toxin, its photo‐isomerization alters the protein distribution on liquid‐ordered domains and induces submicrometer‐sized protein lakes. This may offer new strategies for controlling the structure of biological membranes including membrane‐interacting proteins.
Introduction
The plasma membrane of eukaryotic cells is a complex entity with many different lipids and proteins segregating into various domains.[ 1 , 2 ] Lipid segregation plays a crucial physiological role in mammalian membranes, as these dynamic assemblies are implicated in membrane signaling and trafficking. The outer leaflet of the plasma membrane comprises phospholipids, sphingolipids, sphingomyelins, and glycosphingolipids, as well as cholesterol. Mixing these lipids in vitro results in liquid‐liquid phase separated membranes with a liquid‐ordered phase enriched in sphingolipids and cholesterol and a liquid‐disordered phase enriched in fluid‐phase lipids.[ 3 , 4 , 5 ] In mammalian cells, these more ordered phases are controversially discussed in terms of ‘lipid rafts’, which might contribute to the membrane lateral organization[ 6 , 7 ] and can segregate particular lipids and proteins in the plane of the membrane.[ 8 , 9 ] However, observing these raft domains in cellular membranes remains still a challenge owing to their size, their dynamics, and the overall complexity of the plasma membrane.[ 10 , 11 ]
Thus, a large number of studies focused on artificial bilayers composed of lipid mixtures resembling the outer leaflet of mammalian plasma membranes. They have the advantage that the chemical composition can be precisely controlled. Moreover, synthetic and non‐natural lipids can be readily inserted serving as nano‐tools to investigate and control membrane properties.[ 12 , 13 ] Among them, photo‐switchable lipids appear to be particularly interesting. [14] These photo‐lipids harbor a light‐sensitive azobenzene moiety that isomerizes from the trans‐ to the cis‐configuration upon UV light irradiation [15] changing the geometry of the photo‐lipids from a cylindrical to a more conical shape. This geometrical change has been shown to influence the fluidity of supported membranes, [16] the permeability of vesicles,[ 17 , 18 , 19 ] as well as their morphology, [20] and to induce membrane fusion. [21] They have also been shown to trigger the demixing of the l o phase and the rearrangement of lipid domains in phase‐separated bilayers.[ 22 , 23 , 24 ]
The latter characteristic of photo‐lipids can be exploited to manipulate not only the phase behavior of the membrane itself but also protein‐lipid interactions that are known to occur at ordered domains, which are enriched in sphingolipid receptors. Several sphingolipids specifically serve as docking sites for viral and bacterial toxins. E. g., the cellular uptake of the viruses SV40 or HIV‐1 is mediated by protein‐glycosphingolipid interactions.[ 25 , 26 ] Cholera toxin B binds to GM1, [27] whereas Shiga toxin B binds specifically to Gb3 in phase‐separated membranes.[ 28 , 29 ] Hence, controlling the lateral organization of specific glycosphingolipids serving as receptors for bacterial toxins can become valuable to elucidate the requirements for the protein to bind. In the present work, we focused on the glycosphingolipid Gb3 serving as a receptor for Shiga toxin (STx) that is located in the plasma membrane of the host cell. STx is a cytotoxic protein produced by Shigella dysenteriae and E. coli. [30] Clustering of STx upon plasma membrane binding has been suggested to be an essential step in the formation of membrane invaginations which lead to the internalization of the toxin into the host cells.[ 31 , 32 ] In artificial membrane systems, the B subunits of Shiga toxin (STxB) have been shown to bind only to the l o phase of liquid‐liquid phase‐separated membranes indicating that Gb3 is recruited to and/or localized in the l o domains.[ 33 , 34 , 35 ]
To control the partitioning of Gb3 in phase‐separated membranes and to investigate the influence on lipid segregation and protein binding, photo‐Gb3 glycosphingolipids were synthesized. A photo‐switchable azobenzene group was inserted at four different positions of the fatty acid so that it was either close to the headgroup or on the opposite side. We investigated domain reorganization as well as protein binding as a function of photo‐Gb3 concentration and position of the azobenzene group in phase‐separated supported lipid membranes using confocal fluorescence microscopy and atomic force microscopy.
Results and Discussion
Synthesis and characterization of photo‐Gb3s
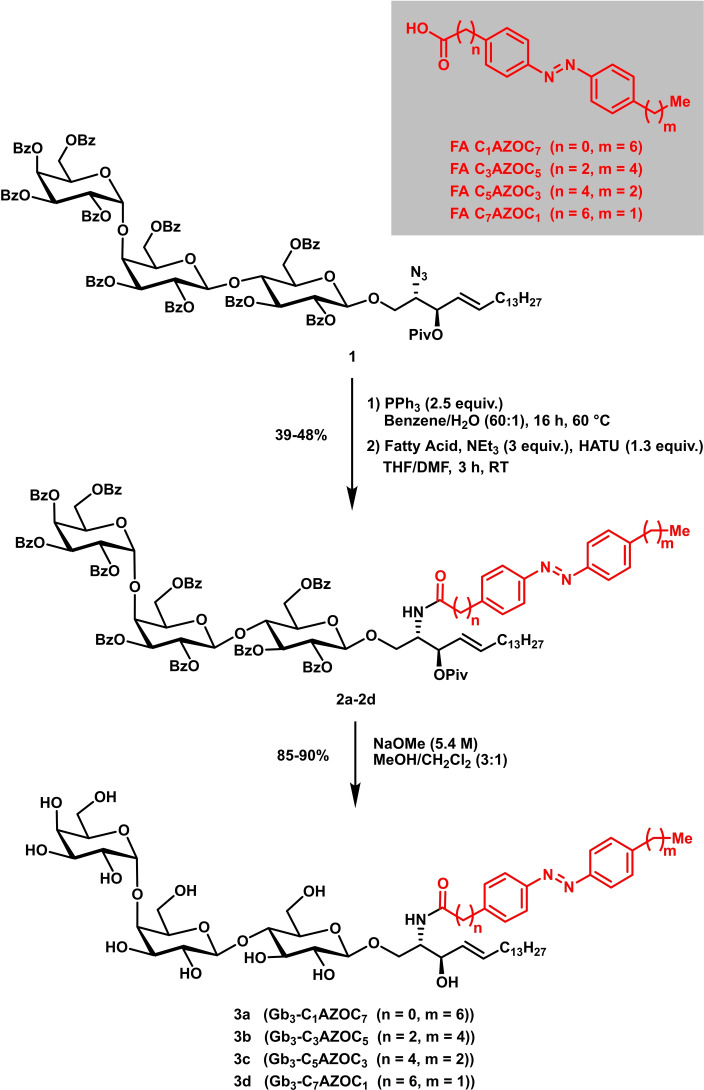
First of all, four different Gb3 glycosphingolipids with different photo‐switchable fatty acids (FA) were synthesized. The synthesis of the respective glycosyl building blocks, the assembly of the trisaccharidic head group, and the attachment of the sphingosine alcohol with a latent amine group (protected as azide moiety) have been described before by our laboratories.[ 36 , 37 , 38 ] Thus, compound 1 was converted using Staudinger conditions to the respective amine that was coupled using common coupling peptide conditions (HATU, NEt3) with azobenzene‐containing fatty acids to afford compounds 2 a–2 d in yields ranging from 39–48 % (Figure 1). The respective fatty acids were synthesized according to a procedure by Trauner and co‐workers. [39] To reach the amphiphilic photo‐switchable glycosphingolipids deprotection of benzoate and pivaloate esters using Zemplén conditions took place affording the desired compounds in yields ranging from 85–90 % (3 a–3 d).
Figure 1.
Synthetic route to four different photo‐switchable Gb3 glycosphingolipids (photo‐Gb3s).
The thermal relaxation of the photo‐Gb3 glycosphingolipids was investigated in phase‐separated lipid vesicles (Figure S17 and Table S1, Supporting Information). After irradiation at 365 nm for 3 min to ensure that all photo‐Gb3 molecules are in cis‐configuration, the absorbance at 340 nm was measured over time. The increase in absorbance reports on the transition to the trans‐configuration. The lifetime of the cis‐configuration of all photo‐Gb3 molecules was in the range of about 10 h indicating that they are stable for a long time period in agreement with previous studies on photo‐lipids.[ 15 , 40 ]
Influence of photo‐Gb3 lipids on membrane domain organization
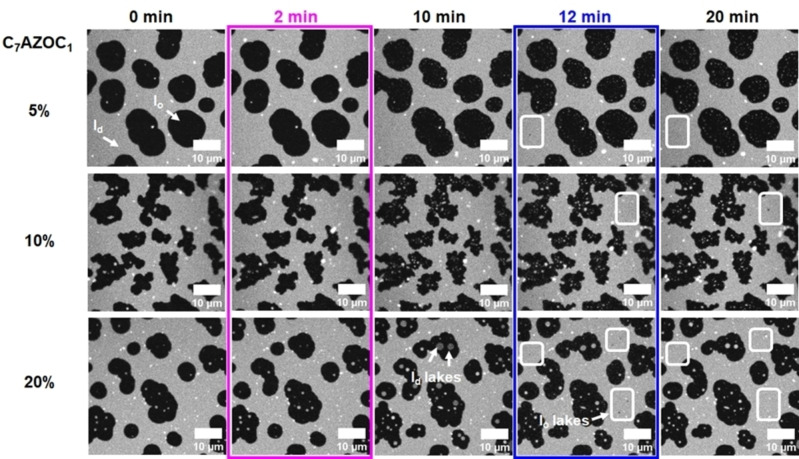
To investigate the influence of the photo‐Gb3 lipids on coexisting liquid‐ordered (l o)/liquid‐disordered (l d) membranes, first supported lipid bilayers (SLBs) composed of DOPC/sphingomyelin (SM18)/cholesterol with increasing concentrations of the photo‐Gb3 C7AZOC1 were prepared on mica surfaces. Fluorescence micrographs show that the lipid dye ATTO 655 DOPE partitions into the l d phase as expected (Figure 2, 0 min). The l o domains appear black. Upon UV irradiation (Figure 2, 2 min), small bright domains (termed “l d lakes” [39] ) appear in the l o domains that become visible in the fluorescence micrographs after some minutes (Figure 2, 10 min; Movie S1, Supporting Information). Concomitantly, the individual l o domains, in which the l d lakes appear, grow in size. The fraction and size of the l d lakes are a function of the Gb3‐C7AZOC1 concentration in the lipid mixture, indicating that they are directly related to the trans‐cis isomerization of the azobenzene moiety of the Gb3‐C7AZOC1 compound (Figure 2, 10 min).
Figure 2.
Domain reorganization of phase‐separated membranes containing increasing concentrations of Gb3‐C7AZOC1. SLBs were composed of DOPC/SM18/cholesterol/Gb3‐C7AZOC1/ATTO 655 DOPE (5 %: 39 : 35 : 20 : 5 : 1, 10 %: 37 : 30 : 22 : 10 : 1, 20 %: 37 : 20 : 22 : 20 : 1). ATTO 655 DOPE partitions into the l d phase (bright), while the l o domains appear black. The membrane was illuminated with UV light after 2 minutes (magenta box) and blue light after 12 minutes (blue box). Upon UV light irradiation, l d domains (“l d lakes”) appear within the l o domains (images at 10 min). Blue light irradiation induced the formation of small l o domains (“l o lakes”) in the l d phase (white squares, images at 12 and 20 min).
While these l d lakes are resolved in mixtures containing 20 mol % Gb3‐C7AZOC1, they are less prominent in presence of 10 mol % and even not resolved in the fluorescence micrographs of mixtures with 5 mol %. As ATTO 655 DOPE partitions into the l d lakes, we assume that these membrane domains are less ordered. A similar result with photo‐ceramides in coexisting l o/l d membranes was reported by Frank et al. [39] As we assumed that also in membranes containing 5 mol % Gb3‐C7AZOC1 l d lakes are formed, we took atomic force micrographs (Figure S18A–C, Supporting Information). Indeed, with this high‐resolution technique, small l d lakes became discernable within the l o domains upon UV irradiation (Figure S18D, Supporting Information).
Upon blue light irradiation (Figure 2, 12 min), the fraction and size of l d lakes get again reduced within the l o domains (Figure 2, 20 min; Movie S1, Supporting Information) suggesting that the l d phase marker re‐partitions from the l d lakes into the surrounding l d phase. This re‐partitioning i. e., the disappearance of the small l d lakes upon blue light irradiation was also verified in lipid mixtures containing 5 mol % Gb3‐C7AZOC1 by atomic force micrographs (Figure S18D, Supporting Information). The process of inducing (UV light) and dispersing (blue light) the l d lakes was reversible and could be performed several times on one membrane (Movie S2, Supporting Information). Moreover, upon blue light irradiation, very small l o domains (termed “l o lakes”) became observable in the l d phase. These small l o lakes appear to be much more mobile than the l d lakes owing to their small size.[ 41 , 42 ]
To ensure that the observed domain formation is not induced by the irradiation of the lipid mixture itself, we performed control experiments with phase‐separated lipid mixtures in the absence of the photo‐Gb3. No changes in domain morphology were observed in the fluorescence micrographs (Figure S19, Movie S3, Supporting Information) and in the atomic force micrographs (Figure S20, Movie S4, Supporting Information) upon irradiation with UV and blue light, confirming that the integrity of the membrane was not affected by the irradiation process. We also performed control experiments on membranes in the presence of photo‐Gb3 and the fluorescent dye ATTO 655 DOPE to ensure that the fluorescence excitation does not influence the morphology of the photo‐Gb3‐containing bilayers (Figures S21 and S22, Supporting Information). In none of the controls, a change in membrane morphology was observed. Thus, we conclude that the observed domain appearance is attributed to photo‐Gb3 isomerization.
Domain organization as a function of the azobenzene‐group position in the fatty acid
The azobenzene moiety in the fatty acid in the trans‐ or cis‐configuration greatly alters the packing parameter of the Gb3 lipid. This influences the partitioning of Gb3 in the l o or l d phase changing the entire domain organization. To investigate how the position of the azobenzene group within the fatty acid influences the domain organization, we synthesized four different photo‐Gb3 lipids with the azobenzene group at different positions within the fatty acid. To observe a large effect of domain organization we added 20 mol % of the corresponding photo‐Gb3 in the lipid mixture and performed the irradiation cycle as described in Figure 2.
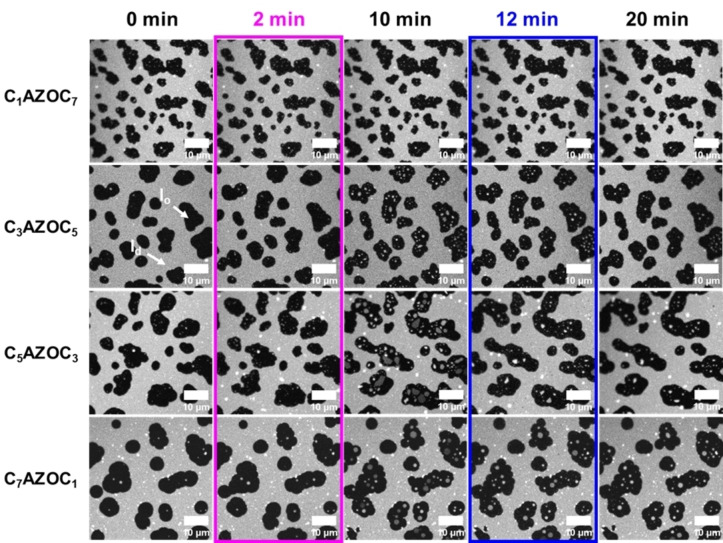
The fluorescence micrographs are shown in Figure 3. The results demonstrate that UV irradiation of SLBs containing Gb3‐C7AZOC1 and Gb3‐C5AZOC3 leads to l d lakes that are very prominent in the l o domains, while the l d lakes are significantly smaller in the case of Gb3‐C3AZOC5. For Gb3‐C1AZOC7 UV irradiation does not even significantly alter the domain organization. Irradiation with blue light again reduces the fraction and size of the l d lakes if they were induced before. The l o lakes occur very seldom in the l d phase and are very transient.
Figure 3.
Impact of the position of the azobenzene group on domain reorganization. Fluorescence micrographs of SLBs composed of DOPC/SM18/cholesterol/photo‐Gb3/ATTO 655 DOPE, 37 : 20 : 22 : 20 : 1. The l o phase appears black while the l d phase is labeled with ATTO 655 DOPE. The membrane was illuminated with UV light after 2 minutes (magenta box) and blue light after 12 minutes (blue box).
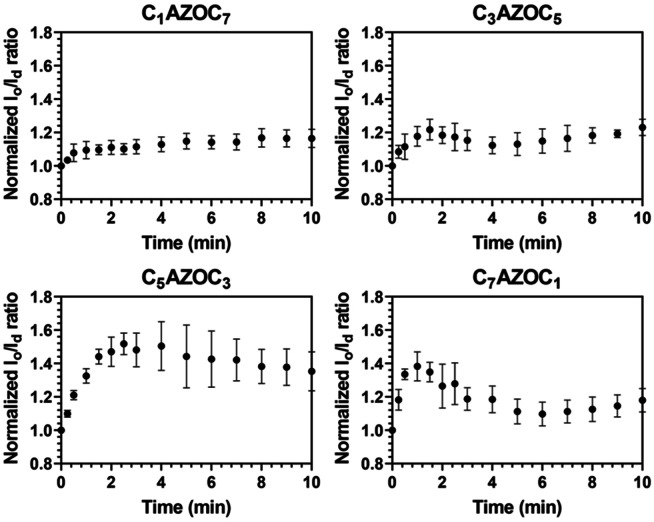
To quantify the influence of the azobenzene position on the fraction of l o to l d phase upon UV irradiation, the l o/l d area ratio was calculated as a function of time after UV irradiation (Figure 4). As the cis‐isomerization of the photo‐Gb3 leads to recruitment of l d phase lipids into the l o domains, two effects intermingle. As long as the l d lakes are not resolved in the l o domains in the fluorescence micrographs, the l o domains appear to grow in size leading to an increase in the l o/l d ratio. If the l d lakes become discernable, which occurs about 1–2 min after UV irradiation, the l o/l d ratio starts decreasing. Dependent on the position of the azobenzene moiety in the fatty acid side chain, the decrease is more or less pronounced after 10 min.
Figure 4.
Influence of the position of the azobenzene group in the fatty acid of the photo‐Gb3 on the l o/l d ratio upon UV irradiation at t=0 min. SLBs were composed of DOPC/SM18/cholesterol/photo‐Gb3/ATTO 655 DOPE, 37 : 20 : 22 : 20 : 1 (n/n). The error bars represent the standard deviation of the mean of 3–4 independent experiments.
Upon cis‐isomerization, photo‐lipids are supposed to adopt a more conical shape, as the azobenzene moiety requires more space [43] resulting in an increased mean molecular area [17] and a reduced chain length. [16] Thus, they more likely partition in the fluid phase. However, in the trans‐configuration they can partition in the l o phase dependent on the position of the azobenzene group. [39] In the case of Gb3‐C1AZOC7, the azobenzene group is close to the headgroup which, in addition to steric constraints, might interfere with the hydrogen bonding between the ceramide and the sphingomyelin in the headgroup region. These effects might decrease its partitioning into the l o domains and hence its ability to recruit l d phase lipids into the l o domains upon photoisomerization. Another aspect to be considered is the efficiency of photoisomerization of Gb3‐C1AZOC7. We observed the formation of l d lakes only after 6 min (Movie S5, Supporting Information). In contrast, in the case of Gb3‐C7AZOC1 the azobenzene group is far away from the headgroup and its geometry allows a better packing with l o phase lipids.
Upon blue light irradiation, the fraction and size of the l d lakes get reduced if they have been formed upon UV light irradiation (Figure S23, Supporting Information). In addition, in presence of Gb3‐C7AZOC1 and Gb3‐C5AZOC3, a large number of small l o lakes became visible in the l d phase immediately after blue light irradiation and they remained for about 3 min. However, in the presence of Gb3‐C1AZOC7 and Gb3‐C3AZOC5, only very few or no l o lakes were observed.
Even though we observed l o lakes upon blue light irradiation, their quantification was not as straightforward as for the l d lakes owing to their small size and their transient appearance. To compare the four different photo‐Gb3 derivatives, we determined the change in l o phase area as Δ(l o/l d) 40 min after blue light irradiations (Table 1).
Table 1.
Influence of the position of the azobenzene group on membrane ordering. Δ(l o/l d) reports on the change in l o/l d ratio at t=40 min after blue light irradiations. Membranes were composed of DOPC/SM18/cholesterol/photo‐Gb3/ATTO 655 DOPE, 37 : 20 : 22 : 20 : 1 (n/n). Before blue light irradiation, the samples were UV irradiated (t=0 min). The errors are the standard deviation of the mean of 2–3 independent experiments.
|
|
C1AZOC7 |
C3AZOC5 |
C5AZOC3 |
C7AZOC1 |
|---|---|---|---|---|
|
Δ(l o/l d) |
0.01±0.01 |
0.07±0.03 |
0.08±0.01 |
0.11±0.02 |
The result indicates that for an ordering of the membrane to occur, the azobenzene moiety needs to be far away from the lipid headgroup. These results are in agreement with the idea that the Gb3 molecules with the azobenzene group close to the headgroup partition preferentially in the l d phase also in the trans‐configuration as proposed for photo‐ceramides. [39] If this is the case the cis‐photoisomerization does not alter the phase behavior of the membrane. Only if the photo‐Gb3 partitions into the l o phase in the trans‐configuration, does the photoisomerization to cis recruit l d phase lipids leading to a re‐organization of the membrane. Our results demonstrate that the complex sugar headgroup of Gb3, which is even more complex than a galactose (galactocerebroside) or a phosphocholine group (sphingomyelin) [44] does not interfere with the photoisomerization in the liquid‐liquid phase‐separated membranes. If the azobenzene group is localized at the end of the fatty acid, the trans‐configuration of the Gb3 (dark adapted or blue light illumination state) appears to predominantly localize within l o domains, while the cis‐configuration (UV‐illumination state) induces l d lakes within l o domains thus reorganizing the l o domains and expanding them.
One particular feature of the Gb3 molecule is its ability to serve as a receptor lipid for Shiga toxin, which is known to only bind to Gb3 containing l o domains in artificial liquid‐liquid phase separated membranes. [33] Hence the question arose of how the photoisomerization of the photo‐Gb3 derivatives alters the binding behavior of Shiga toxin B (STxB) and how binding influences membrane domain re‐organization.
Shiga Toxin B interaction with Gb3‐C7AZOC1
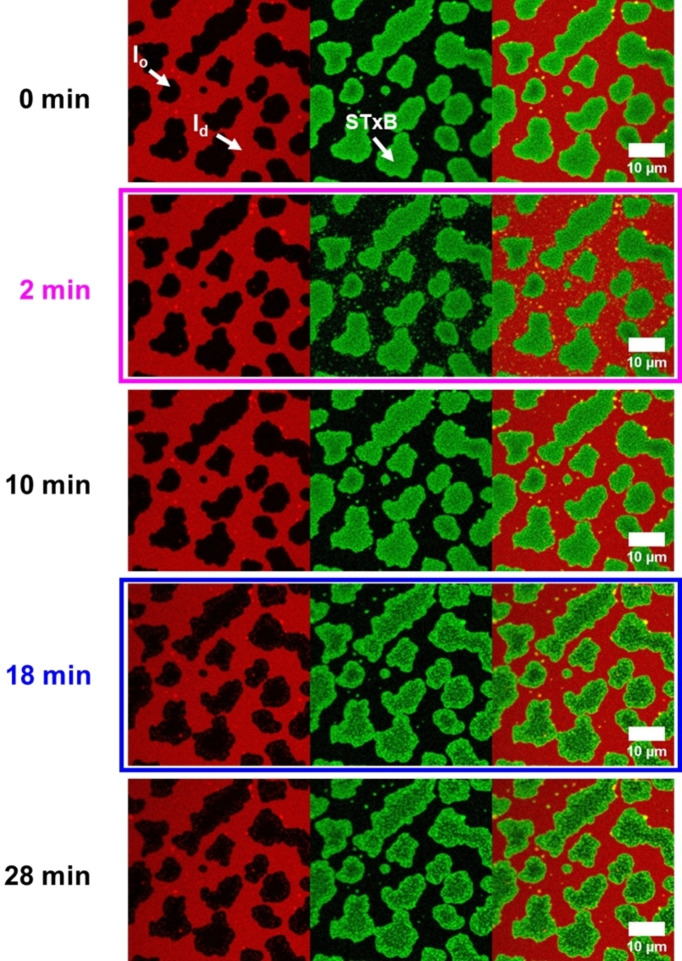
We performed the protein binding studies with Gb3‐C7AZOC1, which we expect to be preferentially localized in the l o phase in trans‐configuration and greatly alters the l o domains upon cis‐isomerization (see Figures 3 and 4). Before UV irradiation, phase‐separated SLBs containing 10 mol % of Gb3‐C7AZOC1 were incubated with Cy3‐labeled STxB. Fluorescence micrographs (Figure 5, 0 min) reveal that STxB binds to the l o phase in agreement with results obtained on non‐modified Gb3 molecules.[ 28 , 29 ] This finding indicates that protein binding is not influenced by the azobenzene moiety at the end of the fatty acid and demonstrates that Gb3 in the trans‐configuration (dark adapted) is mainly localized in the l o phase after STxB binding, despite the relatively bulky azobenzene moiety in the fatty acid. We know from a previous study that the fatty acid attached to Gb3 influences the number of bound Gb3 molecules to the pentameric STxB structure with fewer occupied binding sites in the case of a C24:1 mono‐unsaturated fatty acid compared to a C24:0 saturated one.[ 28 , 29 ] Moreover, the protein density on the membrane is a function of the chemical structure of the fatty acid of the Gb3 molecule resulting in areas of low and high protein density.[ 28 , 29 ] From the Cy3 fluorescence micrographs, we conclude that the protein bound to the photo‐Gb3 is evenly distributed on the surface of the l o domains. As the azobenzene moiety has a planar geometry in the trans‐configuration, [15] it not only allows lipid packing in the l o phase of the phase‐separated membrane, but might also occupy several Gb3 binding sites of STxB resulting in a dense protein packing on the membrane surface.
Figure 5.
Fluorescence micrographs of STxB binding to photo‐Gb3‐C7AZOC1. The planar membrane (red) is composed of DOPC/SM18/cholesterol/C7AZOC1/ATTO 655 DOPE, 37 : 30 : 22 : 10 : 1. 300 nM Cy3‐STxB (green) is added. The membrane was illuminated with UV light after 2 minutes (magenta box) and blue light after 18 minutes (blue box).
To support this notion, we also performed atomic force microscopy on these membranes. A phase‐separated SLB containing 10 mol % of Gb3‐C7AZOC1 was incubated with 300 nM of STxB. Domains are visible that are higher than the surrounding bilayer with an average height difference of 3.0±0.6 nm (Figure 6A/B, Table 2). As we know from the fluorescence micrographs that STxB binds solely to the l o phase, the observed height difference is the difference between the membrane in the l d phase and the membrane in the l o phase (l o domains) with bound STxB. The additional height difference of 1.5 nm compared to that between the l d and l o phase in the absence of bound protein of 1.5±0.5 nm (Figure 6D, Table 2) demonstrates that the protein has been homogeneously bound to the l o phase domains.
Figure 6.
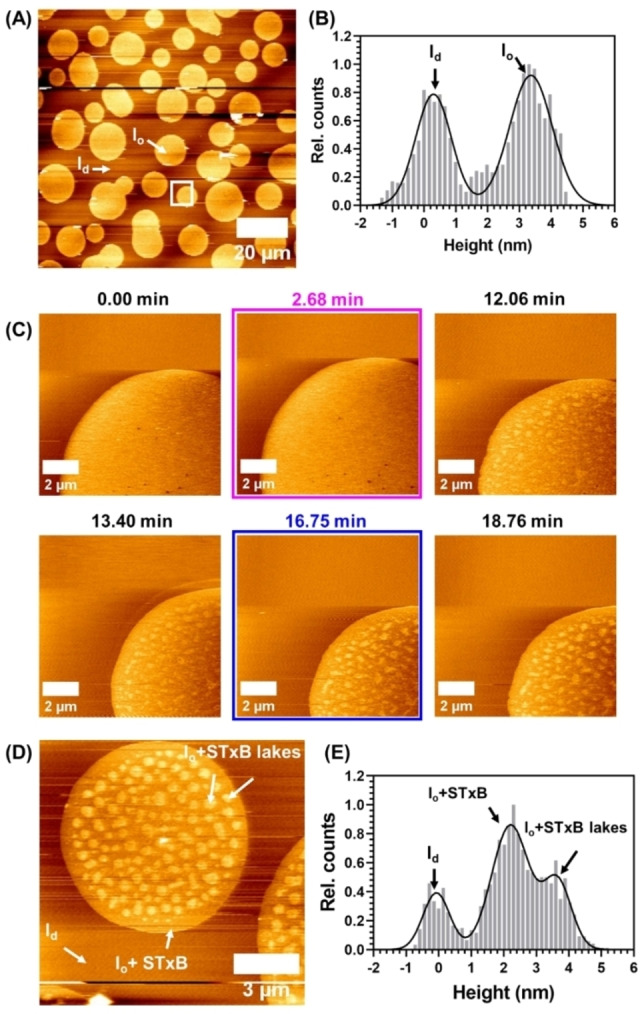
Atomic force micrographs of supported lipid bilayers composed of DOPC/SM18/cholesterol/C7AZOC1/ATTO 655 DOPE, 37 : 30 : 22 : 10 : 1 (n/n) after addition of 300 nM of STxB. The membrane was illuminated with UV light after 2.68 min (magenta box) and blue light after 16.75 minutes (blue box). (A) Overview image of the membrane and (B) histogram analysis (white line) before isomerization. (C) Time series of a zoom in (white square in A) of the membrane. (D) Zoom in on a domain with protein lakes and (E) histogram analysis.
Table 2.
Average height differences were observed between the l d phase, the l o phase, and STxB. The height differences were obtained using the height histograms reported in Figures 6B/E and Figure S20C. The l d phase, which has the lowest height, was taken as a reference in all cases.
|
|
Before irradiation |
After irradiation |
|||
|---|---|---|---|---|---|
|
|
l o‐l d |
(l o+STxB)‐l d |
l o‐l d |
(l o+STxB)‐l d |
(l o+STxB)‐l d(protein lakes) |
|
Δh/nm |
1.5±0.5 |
3.0±0.6 |
1.5±0.5 |
2.3±0.5 |
3.7±0.4 |
No substructures are observed on the l o domains in agreement with the observed homogeneous Cy3 protein fluorescence. Irradiation with UV light did not significantly change the membrane organization (Movie S7, Supporting Information). Even after 10 min (Figure 5, 10 min) the l o and l d phase remain rather constant and the protein is bound to the l o domains. It only appears as if the green fluorescence of the protein becomes grainier over time (compare Figure 5, 0 min, and 10 min). As the Gb3 molecules are bound to the protein, they are not freely moving [45] and remain in the l o domain even though they are in the cis‐configuration. Hence, an exchange of lipids is greatly hampered and probably very slow so that the recruitment of l d phase lipids does not become visible after 10 min as observed for Gb3‐C7AZOC1 in the absence of bound protein (see Figures 2 and 3). To obtain a higher resolution on the photoisomerization process in the presence of bound STxB, we performed atomic force microscopy (Figure 6C). After UV irradiation, clusters with an additional height difference appeared. However, they only become visible about 8–10 min after UV irradiation (Movie S6, Supporting Information).
This result supports the notion that the Gb3‐induced reorganization upon trans‐cis isomerization is very slow because the Gb3 molecules are bound underneath the protein. However, there is a reorganization process that leads to a substructure on the l o domains, which is identified by two different height differences (Figure 6 D/E, Table 2). This substructure was found in all l o domains (Figure S24A, Supporting Information). We refer to the higher structure of 3.7±0.4 nm (Table 2) with respect to the l d phase as “protein lakes”. These higher structures were stable for the entire recording time and did not disperse upon blue light irradiation. From the atomic force micrographs, we determined the size of these protein lakes to be 10–500 nm (Figure S24B).
A corresponding fluorescence z‐stack was recorded demonstrating that the green protein fluorescence is inhomogeneously distributed on the l o domains (Figure S25). From the high fluorescence intensity and the height of 3.7 nm (Table 2), we conclude that STxB is bound to Gb3 with high density on l o phase lipids leading to protein lakes, while less dense protein is bound in the surrounding area on the l o domain (green fluorescence is detectable on the entire l o domains) with probably more l d phase lipids underneath the protein (height difference of only 2.3 nm, Table 2) as a result of the Gb3 in the cis‐configuration.
Recently Glaubitz and co‐workers have shown by NMR spectroscopy that the isomerization triggers an overall reduction in the acyl chain order parameter S CH in close proximity to the azobenzene switch. [46] Thus, the switch to the cis‐configuration apparently produces two populations of bound STxB. As the lipid diffusion is very slow, the process cannot be readily reversed upon blue light irradiation.
These results show that the azobenzene switch not only alters the fluidity of a membrane [16] but also impacts the interaction with a protein. While we are not aware of any study investigating the influence of a trans‐cis photoisomerization on the binding behavior of a peripherally bound protein, Glaubitz and coworkers reported on the influence of the trans‐cis isomerization of azo‐PC on the mobility of the membrane protein DgkA embedded in the lipid bilayer. [46] In general, trans‐cis photoisomerization provides a simple non‐invasive method to alter the phase behavior of membranes that influences the partitioning of membrane proteins similar to what can be observed if the temperature is changed [47] or lipids are uncaged. [45]
Conclusion
The synthesis of a new family of photo‐Gb3 glycosphingolipids differing in the position of the light‐sensitive azobenzene group in the Gb3 fatty acid enabled us not only to switch the membrane morphology of liquid‐liquid phase separated supported lipid bilayers by light, but also to modulate the lateral organization of bound Shiga toxin B that specifically interacts with the receptor lipid Gb3. We conclude that the Gb3‐compounds induced lipid domain rearrangements with the largest influence of the trans‐cis photoisomerization, if the azobenzene is positioned at the end of the fatty acid. Such light switchable receptor lipids may offer new strategies for controlling the morphology of biological membranes as well as the binding properties of membrane‐interacting proteins. Specifically, one can envision to alter the membrane binding mode of Shiga toxin by light, thus influencing the internalization process of the protein into the cells. In this context, it would be desirable to shift the absorption maximum of the light‐switching units to larger wavelengths to reduce the energy input and better access living cells and tissue.
Experimental section
Materials: 1,2‐Dioleoyl‐sn‐glycero‐3‐phosphocholine (DOPC), N‐stearoyl‐d‐erythro‐sphingosyl‐phosphorylcholine (SM18), cholesterol and BODIPY‐cholesterol were purchased from Avanti Polar Lipids (Alabaster, AL, USA) and ATTO 655 DOPE from ATTO Technology (Siegen, Germany). V1‐Mica was obtained from Science Services (Munich, Germany). Petri dishes were purchased from Mattek (Ashland, MA, USA). Tris(hydroxymethyl)aminomethane buffer (Tris) was prepared as followed: 20 mM Tris, 100 mM NaCl, and 1 mM CaCl2. Phosphate buffered saline (PBS) was prepared using 136 mM NaCl, 2.7 mM KCl, 8.1 mM Na2HPO4 and 1.5 mM KH2PO4. All buffers were adjusted to pH 7.4 and filtered with Sartorius 0.2 μm RC membrane filters (Göttingen, Germany).
Chemical synthesis of photo‐Gb3s: For detailed information, see the Supporting Information.
Purification of Shiga Toxin B: The expression of STxB was carried out following a protocol published before [38] with small modifications. In brief, STxB was expressed in E. coli (T7 Express, New England Biolabs, Frankfurt, Germany) in a shaker flask culture (10 times 200 ml). At an optical density of 0.5, protein expression was induced by 0.5 mM IPTG. Cells were cultured at 30 °C overnight and harvested after 30 min centrifugation (15 000×g at 4 °C). The cell pellet was suspended in 20 mM Tris buffer (500 mM NaCl, 1 mM EDTA, 1 % Triton, pH 8.5) for 30 min at 4 °C and subsequently lysed using a French Press. Protein was purified on a chitin column (30 ml) which was equilibrated with Tris buffer (20 mM Tris, 0.5 M NaCl, 1 mM EDTA, pH 8.5) and eluted using the same buffer with an additional 50 mM DTT. The eluate was dialyzed against standard PBS and concentrated using Amicons (3 kDa) to a final concentration of 1.75 mg/ml. The protein was then labeled with Cyanine 3 (Cy3) using a fast conjugation kit (ab188287) from Abcam (Berlin, Germany).
Preparation of phase‐separated lipid vesicles: Lipid mixtures were composed of DOPC/SM18/cholesterol/photo‐Gb3/ATTO 655 DOPE with the following molar ratios: 39 : 35 : 20 : 5 : 1, 37 : 30 : 22 : 10 : 1, and 37 : 20 : 22 : 20 : 1. The mixtures were deposited in glass vials and the solvent evaporated in a water bath at 65 °C using a nitrogen stream. After drying in a vacuum oven for 1 h at 65 °C, the films were hydrated with Tris buffer for 30 min at 65 °C. Samples were vortexed 3 times during 1 min to obtain multilamellar vesicles. In between agitation cycles, samples were incubated 5 min at 65 °C. Small unilamellar vesicles (SUVs) were then prepared by extrusion with a mini‐extruder (Avanti Polar Lipids, Alabaster, AL, USA). 41 passages through a 50 nm polycarbonate membrane were performed. The temperature was maintained at 65 °C using a heating block. Lipid concentration was 0.4 mg/mL and dynamic light scattering measurements (size distribution by intensity) showed vesicles with an average diameter of 100 nm with a unimodal distribution.
Preparation of supported lipid bilayers (SLBs): To obtain SLBs, freshly cleaved mica was glued in 35 mm dishes. The dishes were kept in an oven at 65 °C for 15 min and the vesicles were deposited on the mica in presence of 10 mM CaCl2. After 15 min of incubation at 65 °C, Tris buffer was added to lower the CaCl2 concentration to 1 mM. After 1 h, Tris buffer was replaced with PBS. Prior to observation, samples were slowly cooled down in the dark overnight. For the protein experiments, the samples were incubated under gentle agitation during 1 h with 300 nM of Cy3‐labeled STxB. The samples were then rinsed with PBS to remove unbound protein.
Confocal laser scanning microscopy (CLSM): An LSM 710 CLSM from Carl Zeiss (Jena, Germany) was used to carry out the CLSM experiments. A 633 nm He‐Ne laser (3 % power) and a 561‐10 DPSS laser (3 % power) were used to visualize the ATTO 655 DOPE present in the SLBs and the labeled Cy3‐STxB, respectively. The pinhole was set to 1.5 airy units. Using a W Plan‐Apochromat 63×/1.0 M27 objective, images and time series with 512×512 pixels (16‐bit resolution) were recorded with a pixel dwell time of 1 μs. Images were analyzed with FIJI (http://fiji.sc/Fiji [48] ) and MATLAB Academic version 2017b (The MathWorks, Inc., Natick, MA, USA). For a better visualization, a contrast correction factor (γ) of 0.5 was applied to the images with FIJI.
Atomic force microscopy (AFM): A NanoWizard III device from JPK Instruments (Bruker Nano GmbH, Berlin, Germany) was used to perform the AFM experiments. Biolever mini cantilevers (BL‐AC40TS‐C2) with a spring constant of 0.09 N/m and a resonance frequency of 110 kHz were obtained from Olympus Europa (Hamburg, Germany). Images were recorded in contact mode with a typical 256×256 resolution and a scan rate of 11.4 Hz. Image analysis was performed with Gwyddion SPM software 2.61 (Czech Metrology Institute, http://gwyddion.net/) and JPK data processing software (JPK Instruments). Plots were realized with GraphPad Software 9 (San Diego, CA, USA).
Light irradiation: A pE‐4000 LED from CoolLed Ltd. (Andover, Hampshire, UK) was used to irradiate the samples with UV‐A (365 nm) and blue light (435–460 nm). A Field Max II‐TO power meter from Coherent Inc. (Wilsonville, OR, USA) was used to measure the light intensity (in Watts) and calculate the average irradiance applied to the samples. For the CLSM experiments, the optical fiber of the LED was placed in the collimator at the back of the microscope. The light beam was guided through the objective via the optical path and samples were irradiated for 15 s at a power density of 3 mW/cm2 with UV and 150 mW/cm2 with blue light. For the absorbance spectra, the LED optical fiber was positioned on top of the spectrophotometer. Samples were irradiated for 3 min with UV light at a power density of 3 mW/cm2. For the AFM experiments, the LED optical fiber was fixed above the AFM head. Samples were irradiated for 60 s at a power density of 1.6 mW/cm2 and 130 mW/cm2 with UV and blue light, respectively.
Contributions
S.A. synthesized the photo‐Gb3 glycosphingolipids and performed the NMR characterization. M.B. and C.M. expressed and purified Shiga toxin B. L.S. labeled the protein and performed UV/Vis spectroscopy, CLSM and AFM experiments. L.S. analyzed the data. L.S., D.B.W. and C.S. designed the experiments and wrote the manuscript.
Conflict of interest
The authors declare no competing interest.
1.
Supporting information
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Acknowledgments
The authors thank the Max‐Planck “School Matter to Life” (BMBF) for financial support as well as Jutta Gerber‐Nolte for technical assistance and Daniel Huster for support during protein purification. L.S thanks Claudia Geisler (Institut für Nanophotonik, Göttingen) for advice on the power meter measurements. Open Access funding enabled and organized by Projekt DEAL.
In memory of our late colleague Ulf Diederichsen.
Socrier L., Ahadi S., Bosse M., Montag C., Werz D. B., Steinem C., Chem. Eur. J. 2023, 29, e202202766.
Contributor Information
Prof. Dr. Daniel B. Werz, Email: daniel.werz@chemie.uni-freiburg.de.
Prof. Dr. Claudia Steinem, Email: csteine@gwdg.de.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
- 1. van Meer G., Voelker D. R., Feigenson G. W., Nat. Rev. Mol. Cell Biol. 2008, 9, 112–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sprong H., Van Der Sluijs P., Van Meer G., Nat. Mol. Cell Biol. 2001, 2, 504–513. [DOI] [PubMed] [Google Scholar]
- 3. Veatch S. L., Keller S. L., Biophys. J. 2003, 85, 3074–3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Veatch S. L., Keller S. L., Phys. Rev. Lett. 2005, 94, 3–6. [Google Scholar]
- 5. Veatch S. L., Keller S. L., Phys. Rev. Lett. 2002, 89, 1–4. [Google Scholar]
- 6. Van Meer G., Annu. Rev. Cell Dev. Biol. 1989, 5, 247–275. [DOI] [PubMed] [Google Scholar]
- 7. Levental I., Levental K. R., Heberle F. A., Trends Cell Biol. 2020, 30, 341–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lingwood D., Simons K., Science 2010, 327, 46–50. [DOI] [PubMed] [Google Scholar]
- 9. Simons K., Ikonen E., Nature 1997, 387, 569–572. [DOI] [PubMed] [Google Scholar]
- 10. Munro S., Cell 2003, 115, 377–388. [DOI] [PubMed] [Google Scholar]
- 11. Sezgin E., Levental I., Mayor S., Eggeling C., Nat. Rev. Mol. Cell Biol. 2017, 18, 361–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Laguerre A., Schultz C., Curr. Opin. Cell Biol. 2018, 53, 97–104. [DOI] [PubMed] [Google Scholar]
- 13. Flores J., White B. M., Brea R. J., Baskin J. M., Devaraj N. K., Chem. Soc. Rev. 2020, 49, 4602–4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Morstein J., Impastato A. C., Trauner D., ChemBioChem 2020, 22, 73–83. [DOI] [PubMed] [Google Scholar]
- 15. Bandara H. M. D., Burdette S. C., Chem. Soc. Rev. 2012, 41, 1809–1825. [DOI] [PubMed] [Google Scholar]
- 16. Urban P., Pritzl S. D., Ober M. F., Dirscherl C. F., Pernpeintner C., Konrad D. B., Frank J. A., Trauner D., Nickel B., Lohmueller T., Langmuir 2020, 36, 2629–2634. [DOI] [PubMed] [Google Scholar]
- 17. Pritzl S. D., Urban P., Prasselsperger A., Konrad D. B., Frank J. A., Trauner D., Lohmüller T., Langmuir 2020, 36, 13509–13515. [DOI] [PubMed] [Google Scholar]
- 18. Chander N., Morstein J., Bolten J. S., Shemet A., Cullis P. R., Trauner D., Witzigmann D., Small 2021, 17, 1–11. [DOI] [PubMed] [Google Scholar]
- 19. Pritzl S. D., Konrad D. B., Ober M. F., Richter A. F., Frank J. A., Nickel B., Trauner D., Lohmüller T., Langmuir 2022, 38, 385–393. [DOI] [PubMed] [Google Scholar]
- 20. Pernpeintner C., Frank J. A., Urban P., Roeske C. R., Pritzl S. D., Trauner D., Lohmüller T., Langmuir 2017, 33, 4083–4089. [DOI] [PubMed] [Google Scholar]
- 21. Scheidt H. A., Kolocaj K., Konrad D. B., Frank J. A., Trauner D., Langosch D., Huster D., Biochim. Biophys. Acta Biomembr. 2020, 1862, 183438. [DOI] [PubMed] [Google Scholar]
- 22. Yasuhara K., Sasaki Y., Kikuchi J. I., Colloid Polym. Sci. 2008, 286, 1675–1680. [Google Scholar]
- 23. Urban P., Pritzl S. D., Konrad D. B., Frank J. A., Pernpeintner C., Roeske C. R., Trauner D., Lohmüller T., Langmuir 2018, 34, 13368–13374. [DOI] [PubMed] [Google Scholar]
- 24. Kol M., Williams B., Toombs-Ruane H., Franquelim H. G., Korneev S., Schroeer C., Schwille P., Trauner D., Holthuis J. C., Frank J. A., eLife 2019, 8, 1–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ewers H., Römer W., Smith A. E., Bacia K., Dmitrieff S., Chai W., Mancini R., Kartenbeck J., Chambon V., Berland L., Oppenheim A., Schwarzmann G., Feizi T., Schwille P., Sens P., Helenius A., Johannes L., Nat. Cell Biol. 2010, 12, 11–18. [DOI] [PubMed] [Google Scholar]
- 26. Cook D. G., Fantini J., Spitalnik S. L., Gonzalez-Scarano F., Virology 1994, 201, 206–214. [DOI] [PubMed] [Google Scholar]
- 27. Kenworthy A. K., Schmieder S. S., Raghunathan K., Tiwari A., Wang T., Kelly C. V., Lencer W. I., Toxin Rev. 2021, 13, 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Schütte O. M., Ries A., Orth A., Patalag L. J., Römer W., Steinem C., Werz D. B., Chem. Sci. 2014, 5, 3104–3114. [Google Scholar]
- 29. Windschiegl B., Orth A., Römer W., Berland L., Stechmann B., Bassreau P., Johannes L., Steinem C., PLoS One 2009, 4, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bergan J., Dyve Lingelem A. B., Simm R., Skotland T., Sandvig K., Toxicon 2012, 60, 1085–1107. [DOI] [PubMed] [Google Scholar]
- 31. Johannes L., Billet A., Cancer Metastasis Rev. 2020, 39, 375–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Arumugam S., Bassereau P., Essays Biochem. 2015, 57, 109–119. [DOI] [PubMed] [Google Scholar]
- 33. Römer W., Berland L., Chambon V., Gaus K., Windschiegl B., Tenza D., Aly M. R. E., Fraisier V., Florent J. C., Perrais D., Lamaze C., Raposo G., Steinem C., Sens P., Bassereau P., Johannes L., Nature 2007, 450, 670–675. [DOI] [PubMed] [Google Scholar]
- 34. Pezeshkian W., Gao H., Arumugam S., Becken U., Bassereau P., Florent J. C., Ipsen J. H., Johannes L., Shillcock J. C., ACS Nano 2017, 11, 314–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sibold J., Ahadi S., Werz D. B., Steinem C., Eur. Biophys. J. 2020, 50, 109–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sibold J., Kettelhoit K., Vuong L., Liu F., Werz D. B., Steinem C., Angew. Chem. Int. Ed. 2019, 58, 17805–17813; [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew. Chem. 2019, 131, 17969–17977. [Google Scholar]
- 37. Patalag L. J., Sibold J., Schütte O. M., Steinem C., Werz D. B., ChemBioChem 2017, 18, 2171–2178. [DOI] [PubMed] [Google Scholar]
- 38. Bosse M., Sibold J., Scheidt H. A., Patalag L. J., Kettelhoit K., Ries A., Werz D. B., Steinem C., Huster D., Phys. Chem. Chem. Phys. 2019, 21, 15630–15638. [DOI] [PubMed] [Google Scholar]
- 39. Frank J. A., Franquelim H. G., Schwille P., Trauner D., J. Am. Chem. Soc. 2016, 138, 12981–12986. [DOI] [PubMed] [Google Scholar]
- 40. Morstein J., Kol M., Novak A. J. E., Feng S., Khayyo S., Hinnah K., Li-Purcell N., Pan G., Williams B. M., Riezman H., Atilla-Gokcumen G. E., Holthuis J. C. M., Trauner D., ACS Chem. Biol. 2021, 16, 452–456. [DOI] [PubMed] [Google Scholar]
- 41. Saitov A., Akimov S. A., Galimzyanov T. R., Glasnov T., Pohl P., Phys. Rev. Lett. 2020, 124, 108102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Schütte O. M., Mey I., Enderlein J., Savi F., Geil B., Janshoff A., Steinem C., Proc. Natl. Acad. Sci. USA 2017, 114, E6064–E6071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Klaja O., Frank J. A., Trauner D., Bondar A. N., J. Comput. Chem. 2020, 41, 2336–2351. [DOI] [PubMed] [Google Scholar]
- 44. Hartrampf N., Leitao S. M., Winter N., Toombs-Ruane H., Frank J. A., Schwille P., Trauner D., Franquelim H. G., bioRxiv 2021, 2021.10.11.463883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Carter Ramirez D. M., Kim Y. A., Bittman R., Johnston L. J., Soft Matter 2013, 9, 4890–4899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Doroudgar M., Morstein J., Becker-Baldus J., Trauner D., Glaubitz C., J. Am. Chem. Soc. 2021, 143, 9515–9528. [DOI] [PubMed] [Google Scholar]
- 47. Seeger H. M., Bortolotti C. A., Alessandrini A., Facci P., J. Phys. Chem. B 2009, 113, 16654–16659. [DOI] [PubMed] [Google Scholar]
- 48. Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., Tinevez J. Y., White D. J., Hartenstein V., Eliceiri K., Tomancak P., Cardona A., Nat. Methods 2012, 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.