FIGURE 2.
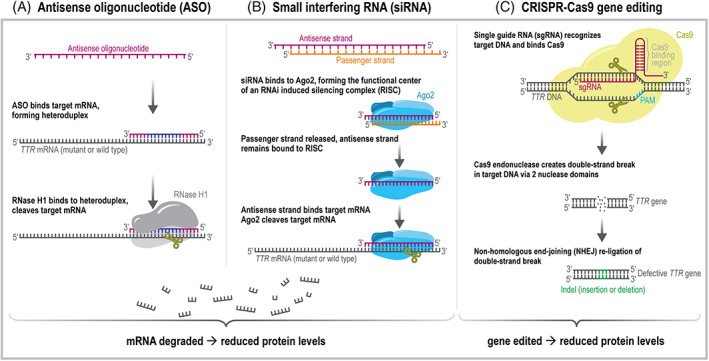
Mechanisms of RNA Silencing and DNA‐Editing Treatments for ATTR. Panel A: Single‐stranded ASOs (eg, inotersen and eplontersen) consist of 10 DNA nucleotides flanked by five chemically modified ribonucleotides. The ASO binds target TTR mRNA, forming a DNA/RNA heteroduplex that binds to and activates ribonuclease H1 (RNase H1). RNase H1 then cleaves the target mRNA. TTR mRNA is degraded, reducing expression of mutant and wild‐type TTR. Panel B: Double‐stranded siRNA (eg, patisiran, vutrisiran) is chemically modified to prevent degradation and is composed of an antisense strand (complementary to the target mRNA) and a passenger strand. The siRNA binds argonaute2 (Ago2) and other proteins to form the RNA‐induced silencing complex (RISC). Once assembled, the passenger strand is released. The antisense strand remains bound to RISC and binds to target TTR mRNA, which is then cleaved by Ago2. TTR mRNA is degraded, reducing expression of mutant and wild‐type TTR. Panel C: CRISPR‐Cas 9 gene editing tool (eg, NTLA‐2001) consists of a single guide RNA (sgRNA) and a Cas9 mRNA sequence (which is translated into Cas9 endonuclease protein once inside the cell). The upstream region of sgRNA contains a 20‐nucleotide sequence that is complementary to TTR DNA and is guided there by a 3‐nucleotide protospacer adjacent motif (PAM). The downstream region of sgRNA binds to Cas9. After cleavage of the TTR DNA via two nuclease domains of Cas9, the double‐strand break is re‐ligated via non‐homologous end‐joining (NHEJ). NHEJ is likely to generate insertion and deletion errors (indels) in the re‐ligated DNA strands, which effectively decreases functional TTR mRNA levels and subsequent protein production.
