Abstract
As one of the most obvious phenotypic traits, the coat color of sheep is an ideal model to study the genetic mechanisms underlying coat color varieties of mammals. One distinguishable coat color is the black-headed type, such as the famous black-headed Dorper sheep from Africa and Bayinbuluke sheep from Asia. In this study, we compared the genome sequences of black-headed and all-white sheep to identify causative genes for the black-headed sheep, including black-headed Dorper vs. white-headed Dorper, as well as Bayinbuluke (black-headed) vs. Small-tailed Han (all-white). The most differentiating region between black-headed sheep and all-white sheep was found to harbor a haplotype covering melanocortin receptor 1 (MC1R) gene. The share of this haplotype by the black-headed sheep from Africa and Asia suggested that the convergent change in the MC1R region is likely to determine this unique coat color. Two missense mutations (g. 14251947T > A and g. 14252090G > A) within this haplotype of MC1R gene were found. We further analyzed whole genome sequence data of 460 worldwide sheep with diverse coat colors and confirmed the association between the MC1R haplotype with pigmentation variations. Our study provides novel insights into coat color genetics in sheep and expands our knowledge of the link between MC1R gene and varying pigmentation patterns in sheep.
Keywords: black-headed sheep, coat color, melanocortin receptor 1, missense mutations, sheep
This study provides new insights into the inheritance of coat color traits in sheep and expands the understanding of the association between the melanocortin receptor 1 gene and different pigmentation patterns in sheep.
Introduction
Coat color is the most distinguishable phenotype of domestic animals as recognized by Darwin (Darwin, 1868). Domestication, adaptation, and artificial selection have produced a large variety of coat colors that become the most noticeable traits of diverse breeds (Cieslak et al., 2011). In modern animal farm animal breeding, the coat color could be used to discriminate between breeds and varieties. Furthermore, coat color becomes an important economic trait for sheep which are raised for skins and wool (Sun et al., 2020). The ancestral coat color of sheep is mainly brown, but domesticated sheep displayed various colors, patterns, and white markings (Lundie, 2011). Therefore, the diverse sheep breeds provide a suitable animal model to study the genetic background of coat color diversity in animals.
Coat color patterns often abide by the Mendelian mode of inheritance and are among the first traits to be studied by a molecular geneticist (Rieder, 2009). Classical candidate genes for the coat color of mammals that have been reported include the agouti signaling protein (ASIP) gene, melanocortin 1 receptor (MC1R) gene, tyrosinase-related protein 1 gene, melanocyte-inducing transcription factor gene, and tyrosine-protein kinase gene. These five genes play key roles in melanin synthesis, differentiation, and migration of melanocytes and thus represent the most widely studied pigmentation genes (Nazari-Ghadikolaei et al., 2018; Yao et al., 2019; Gebreselassie et al., 2020; Jiang et al., 2021; Liang et al., 2021; Patterson et al., 2021). Mutations in the MC1R gene have been found to determine the black phenotype of Chinese sheep (Yang et al., 2013). The black skin and hairs of Masai sheep were also found to be associated with MC1R mutations (Fontanesi et al., 2011).
Of the diverse coat colors in sheep, pigmentation type in head is most distinguishable. The black-headed Dorper from Africa and Bayinbuluke sheep from Asia are two unique breeds that demonstrate similar black-headed patterns where the pigmentation expands from the face to the neck. Meanwhile, a white Dorper breed have also been developed by modern breeding, which could be used as a control to study the black-headed coat color. In this study, we aimed to identify genes and loci affecting pigmentation in head of sheep by analyzing genome sequences of black-headed sheep with other breeds. The results will expand our knowledge of the genetic background and convergent evolution underlying coat color diversities in domestic animals.
Materials and Methods
Ethics statement
All animal experiments were approved by the International Animal Care and Use Committee of the Northwest A&F University (IACUC-NWAFU) in accordance with the regulations of the Administration of Affairs Concerning Experimental Animals of China.
Samples collection, DNA extraction, and SNP calling
Whole-blood samples were collected from the Yulin Shanghe Hu Sheep Breeding Base in black-headed Dorper (n = 10) and white-headed Dorper sheep (n = 5). DNA was extracted using the standard phenol-chloroform method (Köchl et al., 2005). Paired-end sequence data for all individuals were generated using the DNBSEQ T7 platform. The whole genome sequence data generated in this study have been deposited in NCBI SRA database with Project number PRJNA904424. Besides, we collected whole genome sequencing data of 110 individual sheep. Samples were selected to obtain the optimum representation of global sheep diversity, and Supplementary Table S1 provides sample information. All sequences were downloaded from NCBI. After quality filtering of the original FASTQ files, the whole genome sequence was compared with slat (v1.0.3) to obtain the Binary Alignment MAP (.bam) files. Then, slct (1.0.3) was used to construct the gvcf file of each sample and the vcf file was merged by slmgvcf_gpu (v2.0.7) (Zhang et al., 2021). Finally, we used the bcftools-1.13 view module of “QD < 2.0, QUAL < 30.0, SOR > 3.0, FS > 60.0, MQ < 40.0, MQRankSum < -12.5 and ReadPosRankSum <-8.0” to remove SNP sequencing and alignment errors. And we used bcftools-1.13 to filter out variants with a call rate > 80% and minor allele frequency < 0.05 for further analysis.
Population structure and phylogenetic analysis
We used PLINK for principal component analysis (PCA) and ADMIXTURE analysis, and PCA used smartPCA program in EIGENSOFT v5.0 package (Patterson et al., 2006). Population structure analysis was carried out using ADMIXTURE v1.3 (Alexander and Lange, 2011). Neighbor-Joining tree was constructed for the whole-genome SNP, and the MEGA v5.0 (Tamura et al., 2011) and exhibited FigTree v1.4.3 visualization.
Exploration of selective sweep regions
To identify the genomic signatures of selection, we calculated the averaged the fixed index (FST) values (Weir and Cockerham, 1984) across each 10-kb window using the VCFtools v0.1.16 between Bayinbuluke and Small-tailed Han sheep, as well as black-headed Dorper and white-headed Dorper. Besides, we then estimated nucleotide diversity θπ using VCFtools v0.1.16 (--window-pi 10000).To identify the population structure across all sheep populations, we performed a haplotype-based approach across all individuals using ChromoPainter and fineSTRUCTURE to explore haplotype-sharing patterns (Lawson et al., 2012). LDBlockShow software with default parameters was used to reconstructed patterns of linkage disequilibrium between different sheep breeds from a single vcf files (Dong et al., 2021).
Annotation
SnpEff V4.2 annotated SNP based on sheep reference genome Oar_rambouillet_v2.0. It can be divided into intron regions, exonic regions, intergenic regions, splicing sites, upstream, and downstream regions. Those SNPs in exons are further divided into synonymous or non-synonymous SNPs (Lv et al., 2022). Finally, the allele frequencies of SNPs variants in different varieties were calculated by VCFtools v0.1.16, and compared allelic variations of the genes in diverse breeds.
Results
Population structure and relationships
We generated complete genomes from black-headed Dorper (n = 10) and white-headed Dorper (n = 5) with an average sequencing depth of 21.5×. We also downloaded publicly available data from black-headed Dorper (BDP), white-headed Dorper (WDP), Bayinbuluke sheep (BYK), and Small-tailed Han sheep (STH) (supplementary Table S1). In this way, we were able to compare black-headed Dorper (n = 15) and white-headed Dorper (n = 14), Small-tailed Han sheep (n = 23), and Bayinbuluke sheep (n = 34) to identify selection signals associated with black-headed coat color (Figure 1). The Small-tailed Han sheep was used here for comparison with Bayinbuluke sheep as they two shares the same ancestry of Mongolian sheep lineage.
Figure 1.
Experimental design to study the genetics of black-headed sheep.
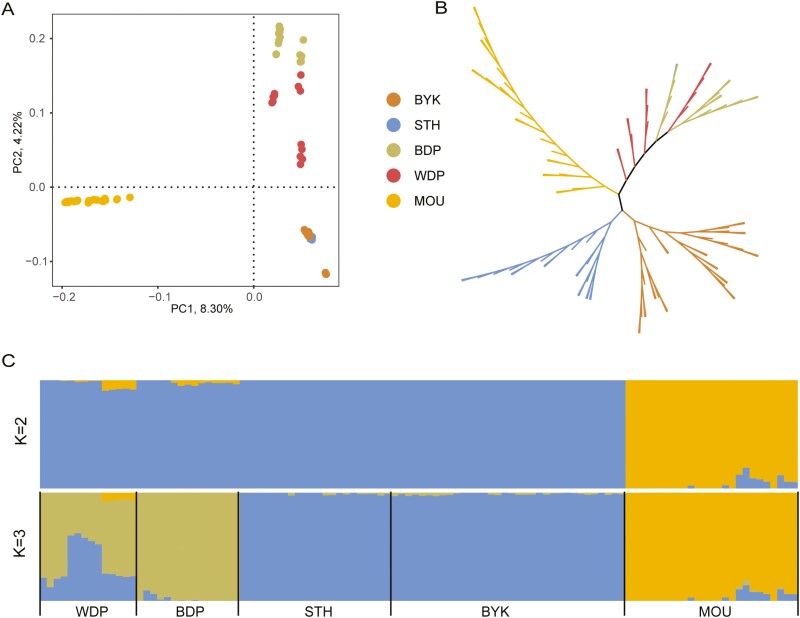
PCA analysis on whole genome sequencing data showed three geographic clusters including African breeds (BDP and WDP), Asian breeds (BYK and STH), and wild (Figure 2A and B). The neighbor-Joining tree generally agrees with the PCA analysis and showed that the black-headed Dorper and white-headed Dorper were mixed in one cluster indicating their relatively low genetic differentiation (Figure 2B). In the ADMIXTURE analysis, when K = 2, the sheep breeds were genetically divided into domestic sheep and Mouflon ancestry; when K = 3, the white-headed Dorper sheep showed clear evidence of shared genome ancestry with black-headed Dorper sheep (Figure 2C).
Figure 2.
Phylogenetic relationships among five sheep populations. (A) PCA of 5 sheep breeds (B)Neighbor-joining tree of the relationships between the five sheep breeds (110 animals).(C) ADMIXTURE results for K = 2 and K = 3, showing low cross-validation error.
Screening genes associated with black-headed sheep
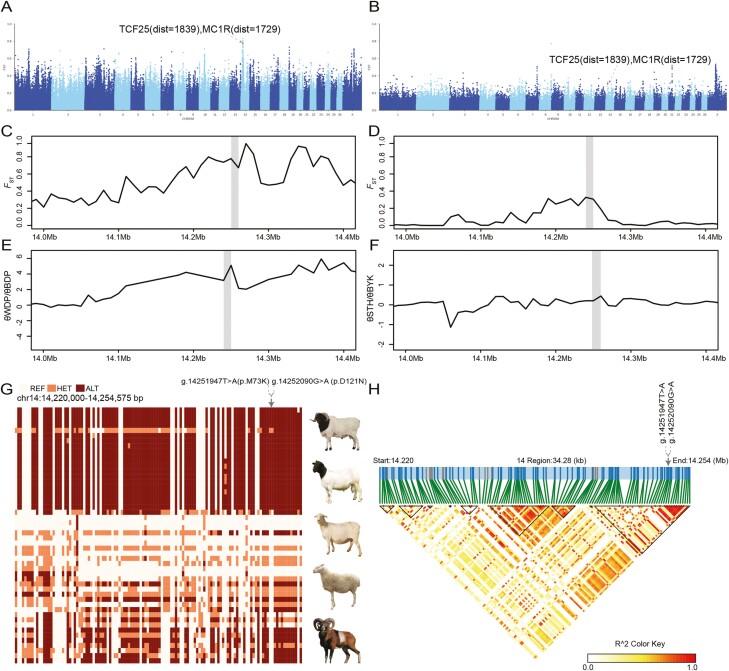
To identify genes related to black-headed sheep, we performed two independent comprisons using the full genome sequences of black-headed Dorper vs. white-headed Dorper, and Bayinbuluke (black-headed sheep) vs. Small-tailed Han (all-white sheep) by calculating the average FST values. The most noticeable signal of FST that segregated in black and white-headed sheep is located in the region of 15.25 to 15.26 Mbp on chromosome 14 (Figure 3A and B). The high FST and low π ratio indicated that this region was subjected to selection in Bayinbuluke and black-headed Dorper sheep (Figure 3C–F). Notably, this region harbors the MC1R gene, which is a well-known genes controlling the switch between eumelanin and phaeomelanin production (Marklund et al., 1996; Bellone, 2010).
Figure 3.
Identification of genes associated with Bayinbuluke and Dorper black-headed traits. (A–B) Manhattan plot of selective sweeps in black-headed Dorper and Bayinbuluke sheep. (C–D) FST at the MC1R gene region. The gray bar spans the MC1R gene model under the strongest selection. (E–F) Nucleotide diversity at the MC1R gene region. (G) SNPs with minor allele frequencies > 0.1 are used to construct haplotype patterns (Chr 14:15.47-15.49 Mb). Two mutations of MC1R gene (g. 14251947T > A and g. 14252090G > A) were both located in the exon region. (H) Haplotype block analysis at the MC1R loci.
Within the selection regions, we found two missense mutations in the protein-coding region of MC1R, namely g.14251947T > A (p.M73K) and g.14252090G > A (p.D121N) residing in the haplotype (Figure 3G). These two variants in MC1R might represent the functional variants affecting the black-headed trait in Bayinbuluke and Dorper sheep. We examined the allele frequencies of two missense mutations (g.14251947T > A and g.14252090G > A) of MC1R gene in many breeds. As these two mutations are linked in one haplotype block (Figure 3H), we mainly analyzed the A allele frequency of g. 14251947T > A. The A allele was highest in Bayinbuluke (BYK, frequency = 64.71%), Sunite (frequency = 62.50%) and black-headed Dorper (BDP, frequency = 100%) (Supplementary Figure S1). In addition, the derived allele A was absent in wild Mouflons, Bighorn sheep, and Argali sheep (Supplementary Figure S1), suggesting that the putative causative mutations emerged after domestication.
MC1R haplotypes in sheep with diverse pigmentation patterns
The classic sheep coat color associated with pigmentation, included black-headed, black-spotted, all-black, all-white, and brown pigment types. We collected 460 resequencing data from 29 sheep breeds each corresponding to one of the five coat colors. Interestingly, our study found that the MC1R gene haplotype indeed had a certain association with different patterns. According to results from haplotype, the white coat color and brown breeds were clustered together, while black-headed, black-spotted, and all-black sheep shared similar haplotypes. As expected, the black-headed Dorper and Bayinbuluke have a unique haplotype (Figure 4). However, it is worth noting that all-black sheep and black spotted sheep also share this cluster with blackheads, suggesting that other loci are also involved to affect the extent and location of melanin deposition, resulting in these different pigmentation patterns.
Figure 4.
Haplotype network based on pairwise differences within MC1R gene. Each circle represents a haplotype, and the size of the circle indicates the number of samples contained in that haplotype. Green represents all-white sheep, yellow for spotted sheep (with black spots on the head or body), blue for black-headed sheep (with black only the head), orange for all-black sheep, and rusty red represents wild Mouflon sheep (brown).
Discussion
Sheep are one of the earliest domesticated animals in the world. It is not only an important source of human meat, but also an important textile material (Rochus et al., 2018). In addition, coat color is an important economic trait for the sheep breeds which are kept for skins and wool, and can also be used for species identification and characterization (Fontanesi et al., 2010). Therefore, it is important to identify genomic regions and genetic variations associated with sheep coat color traits.
We analyzed the genetic basis of the divergence of different head colors in sheep by selective signal and haplotype analysis. Most interestingly, we found the same haplotype in African and Asian sheep with similar black-headed phenotypes, both containing MC1R gene. Black-headed Dorper in African and Bayinbuluke sheep in Asia were raised and selected independently, suggesting that converging changes in the MC1R gene range led to this common phenotype. The MC1R is a seven-transmembrane domain G-protein-coupled receptor, which mainly exists in melanocytes and can be used as a switch to control the synthesis of melanin types for deposition in tissues (Mountjoy et al., 1992; Gebreselassie et al., 2020). When MC1R was activated by melanocyte-stimulating hormone from pituitary, MC1R initiated a downstream signal cascade, resulting in melanin eumelanin production by melanocytes (Swope et al., 2018; Jiang et al., 2021). The MC1R gene was reported to be a potential candidate gene that plays an important role in melanin production and wool pigmentation, and is associated with black fur color in mammals (Våge et al., 1999; Mao et al., 2010; Li et al., 2013; Switonski et al., 2013; Matsumoto et al., 2020). Hepp et al. found that MC1R was involved in the control of dark wool color in Brazilian Creole sheep, where the dominant allele (ED) was only found in colored animals, and the recessive allele (E+) was only homozygous in white individuals (Hepp et al., 2012). In our study, the selective signal and the long haplotype of the selected regions have come from the black-headed sheep breeds. Meanwhile, we also found two missense mutations in the MC1R gene, and hypothesized that the missense mutations may disrupt the melanic pigmentation receptor of MC1R, consequently preventing normal pigmentation of coat color.
A notable example is the conserved role of the MC1R in mammalian pigmentation which could affect the degree and region of melanosis (Andersson, 2003; Yang et al., 2013). We studied the relationship between MC1R haplotypes and coat color using resequencing data from black-headed, black-spotted, all-black, all-white, and Mouflon sheep in 460 sheep. Indeed, we found an obvious haplotype, cluster pattern in which individuals with different degrees of pigmentation clustered together, and white individuals clustered together. Studies have shown that MC1R interacts with other genes to regulate a variety of coat colors, such as ASIP gene (Fontanesi et al., 2011; Hepp et al., 2016). Besides, animal skin pigmentation is a complex trait controlled by multiple genes and may be influenced by different types of multi-gene interactions (Barsh, 1996; Hubbard et al., 2010; Rochus et al., 2019; Shi et al., 2021). Therefore, we speculated that in addition to the influence of MC1R haplotype on black-headed sheep, the coat color of other breeds with black faces, all-black, and black spotted may also be jointly regulated by other coat color genes.
Conclusions
In this study, we found that MC1R is likely to control the black-headed coat color in sheep. We further analyzed MC1R haplotypes using whole genome sequence data available and confirmed the association between MC1R haplotypes and pigmentation patterns in sheep providing novel insights into the coat color genetics of sheep.
Supplementary Material
Acknowledgments
This work was funded by the National Key R&D Program of China (2022YFF1000100 & 2022YFD1300200) and Shaanxi Livestock and Poultry Breeding Double-chain Fusion Key Project (2022GD-TSLD-46-0401). We also thank the National Supercomputing Center in Xi’an for providing computing resources.
Glossary
Abbreviations
- ASIP
agouti signaling protein
- BDP
black-headed Dorper
- BYK
Bayinbuluke
- F ST
fixed index
- MC1R
melanocortin receptor 1
- PCA
principal component analysis
- STH
Small-tailed Han
- WDP
white-headed Dorper
Contributor Information
Qian Zhou, College of Animal Science and Technology, Northwest A&F University, Yangling, Shaanxi 712100, China.
Chunna Cao, College of Animal Science and Technology, Northwest A&F University, Yangling, Shaanxi 712100, China.
Huanhuan Zhang, College of Animal Science and Technology, Northwest A&F University, Yangling, Shaanxi 712100, China.
Yilin Liang, College of Animal Science and Technology, Northwest A&F University, Yangling, Shaanxi 712100, China.
Xinyue Zhang, College of Animal Science and Technology, Northwest A&F University, Yangling, Shaanxi 712100, China.
Yuxin Kang, College of Animal Science and Technology, Northwest A&F University, Yangling, Shaanxi 712100, China.
Wenwen Fang, College of Animal Science and Technology, Northwest A&F University, Yangling, Shaanxi 712100, China.
Xianyong Lan, College of Animal Science and Technology, Northwest A&F University, Yangling, Shaanxi 712100, China.
Ran Li, College of Animal Science and Technology, Northwest A&F University, Yangling, Shaanxi 712100, China.
Chuanying Pan, College of Animal Science and Technology, Northwest A&F University, Yangling, Shaanxi 712100, China.
Conflict of Interest Statement
The authors declare no real or perceived conflicts of interest.
Literature Cited
- Alexander, D. H., and Lange K.. . 2011. Enhancements to the ADMIXTURE algorithm for individual ancestry estimation. BMC Bioinform. 12:246. doi: 10.1186/1471-2105-12-246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson, L. 2003. Melanocortin receptor variants with phenotypic effects in horse, pig, and chicken. Ann. N. Y. Acad. Sci. 994:313–318. doi: 10.1111/j.1749-6632.2003.tb03195.x [DOI] [PubMed] [Google Scholar]
- Barsh, G. S. 1996. The genetics of pigmentation: from fancy genes to complex traits. Trends Genet. 12:299–305. doi: 10.1016/0168-9525(96)10031-7 [DOI] [PubMed] [Google Scholar]
- Bellone, R. R. 2010. Pleiotropic effects of pigmentation genes in horses. Anim. Genet. 41:100–110. doi: 10.1111/j.1365-2052.2010.02116.x [DOI] [PubMed] [Google Scholar]
- Cieslak, M., Reissmann M., Hofreiter M., and Ludwig A.. . 2011. Colours of domestication. Biol. Rev. Camb. Philos. Soc. 86:885–899. doi: 10.1111/j.1469-185X.2011.00177.x [DOI] [PubMed] [Google Scholar]
- Darwin, C. 1868. The variation of animals and plants under domestication. Br. Foreign Med. Chir. Rev. 42:143–166. [PMC free article] [PubMed] [Google Scholar]
- Dong, S. S., He W. M., Ji J. J., Zhang C., Guo Y., and Yang T. L. . 2021. LDBlockShow: a fast and convenient tool for visualizing linkage disequilibrium and haplotype blocks based on variant call format files. Brief. Bioinform. 22:bbaa227. doi: 10.1093/bib/bbaa227 [DOI] [PubMed] [Google Scholar]
- Fontanesi, L., Beretti F., Riggio V., Dall’Olio S., Calascibetta D., Russo V., and Portolano B.. . 2010. Sequence characterization of the melanocortin 1 receptor (MC1R) gene in sheep with different coat colours and identification of the putative e allele at the ovine Extension locus. Small Rumin. Res. 91:200–207. doi: 10.1016/j.smallrumres.2010.03.015 [DOI] [Google Scholar]
- Fontanesi, L., Dall’Olio S., Beretti F., Portolano B., and Russo V.. . 2011. Coat colours in the Massese sheep breed are associated with mutations in the agouti signalling protein (ASIP) and melanocortin 1 receptor (MC1R) genes. Animal. 5:8–17. doi: 10.1017/S1751731110001382 [DOI] [PubMed] [Google Scholar]
- Gebreselassie, G., Liang B., Berihulay H., Islam R., Abied A., Jiang L., Zhao Z., and Ma Y.. . 2020. Genomic mapping identifies two genetic variants in the MC1R gene for coat colour variation in Chinese Tan sheep. PLoS One. 15:e0235426. doi: 10.1371/journal.pone.0235426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepp, D., Gonçalves G. L, Moreira G. R., Freitas T. R., Martins C. T., Weimer T. A., and Passos D. T.. . 2012. Identification of the e allele at the Extension locus (MC1R) in Brazilian Creole sheep and its role in wool color variation. Genet. Mol. Res. 11:2997–3006. doi: 10.4238/2012.May.22.5 [DOI] [PubMed] [Google Scholar]
- Hepp, D., Gonçalves G. L, Moreira G. R., and de Freitas T. R.. . 2016. Epistatic interaction of the melanocortin 1 receptor and agouti signaling protein genes modulates wool color in the Brazilian Creole Sheep. J. Hered. 107:544–552. doi: 10.1093/jhered/esw037 [DOI] [PubMed] [Google Scholar]
- Hubbard, J. K., Uy J. A., Hauber M. E., Hoekstra H. E., and Safran R. J.. . 2010. Vertebrate pigmentation: from underlying genes to adaptive function. Trends Genet. 26:231–239. doi: 10.1016/j.tig.2010.02.002 [DOI] [PubMed] [Google Scholar]
- Jiang, L., Kon T., Chen C., Ichikawa R., Zheng Q., Pei L., Takemura I., Nsobi L. H., Tabata H., Pan H., . et al. 2021. Whole-genome sequencing of endangered Zhoushan cattle suggests its origin and the association of MC1R with black coat colour. Sci. Rep. 11(1):17359. doi: 10.1038/s41598-021-96896-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köchl, S., Niederstätter H., and Parson W.. . 2005. DNA extraction and quantitation of forensic samples using the phenol-chloroform method and real-time PCR. Methods Mol. Biol. 297:13–30. doi: 10.1385/1-59259-867-6:013 [DOI] [PubMed] [Google Scholar]
- Lawson, D. J., Hellenthal G., Myers S., and Falush D.. . 2012. Inference of population structure using dense haplotype data. PLoS Genet. 8:e1002453. doi: 10.1371/journal.pgen.1002453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, M., Tian S., Jin L., Zhou G., Li Y., Zhang Y., Wang T., Yeung C. K., Chen L., Ma J., . et al. 2013. Genomic analyses identify distinct patterns of selection in domesticated pigs and Tibetan wild boars. Nat. Genet. 45:1431–1438. doi: 10.1038/ng.2811 [DOI] [PubMed] [Google Scholar]
- Liang, D., Zhao P., Si J., Fang L., Pairo-Castineira E., Hu X., Xu Q., Hou Y., Gong Y., Liang Z., . et al. 2021. Genomic analysis revealed a convergent evolution of LINE-1 in coat color A case study in water buffaloes (Bubalus bubalis). Mol. Biol. Evol. 38:1122–1136. doi: 10.1093/molbev/msaa279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundie, R. S. 2011. The genetics of colour in fat-tailed sheep: a review. Trop. Anim. Health Prod. 43:1245–1265. doi: 10.1007/s11250-011-9850-0 [DOI] [PubMed] [Google Scholar]
- Lv, F. H., Cao Y. H., Liu G. J., Luo L. Y., Lu R., Liu M. J., Li W. R., Zhou P., Wang X. H., Shen M., . et al. 2022. Whole-genome resequencing of worldwide wild and domestic sheep elucidates genetic diversity, introgression, and agronomically important loci. Mol. Biol. Evol. 39:msab353. doi: 10.1093/molbev/msab353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao, H., Ren J., Ding N., Xiao S., and Huang L.. . 2010. Genetic variation within coat color genes of MC1R and ASIP in Chinese brownish red Tibetan pigs. Anim. Sci. J. 81:630–634. doi: 10.1111/j.1740-0929.2010.00789.x [DOI] [PubMed] [Google Scholar]
- Marklund, L., Moller M. J., Sandberg K., and Andersson L.. . 1996. A missense mutation in the gene for melanocyte-stimulating hormone receptor (MC1R) is associated with the chestnut coat color in horses. Mamm. Genome. 7:895–899. doi: 10.1007/s003359900264 [DOI] [PubMed] [Google Scholar]
- Matsumoto, H., Kojya M., Takamuku H., Kimura S., Kashimura A., Imai S., Yamauchi K., and Ito S.. . 2020. MC1R c.310G>- and c.871G > A determine the coat color of Kumamoto sub-breed of Japanese Brown cattle. Anim. Sci. J. 91:e13367. doi: 10.1111/asj.13367 [DOI] [PubMed] [Google Scholar]
- Mountjoy, K. G., Robbins L. S., Mortrud M. T., and Cone R. D.. . 1992. The cloning of a family of genes that encode the melanocortin receptors. Science. 257:1248–1251. doi: 10.1126/science.1325670 [DOI] [PubMed] [Google Scholar]
- Nazari-Ghadikolaei, A., Mehrabani-Yeganeh H., Miarei-Aashtiani S. R., Staiger E. A., Rashidi A., and Huson H. J.. . 2018. Genome-wide association studies identify candidate genes for coat color and mohair traits in the Iranian Markhoz goat. Front. Genet. 9:105. doi: 10.3389/fgene.2018.00105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson, N., Price A. L., and Reich D.. . 2006. Population structure and Eigenanalysis. PLoS Genet. 2(12):e190. doi: 10.1371/journal.pgen.0020190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson Rosa, L., Martin K., Vierra M., Foster G., Lundquist E., Brooks S. A., and Lafayette C.. . 2021. Two variants of KIT causing white patterning in stock-type horses. J. Hered. 112:447–451. doi: 10.1093/jhered/esab033 [DOI] [PubMed] [Google Scholar]
- Rieder, S. 2009. Molecular tests for coat colours in horses. J. Anim. Breed. Genet. 126:415–424. doi: 10.1111/j.1439-0388.2009.00832.x [DOI] [PubMed] [Google Scholar]
- Rochus, C. M., Tortereau F., Plisson-Petit F., Restoux G., Moreno-Romieux C., Tosser-Klopp G., and Servin B.. . 2018. Revealing the selection history of adaptive loci using genome-wide scans for selection: an example from domestic sheep. BMC Genom. 19:71. doi: 10.1186/s12864-018-4447-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochus, C. M., Westberg Sunesson K., Jonas E., Mikko S., and Johansson A. M.. . 2019. Mutations in ASIP and MC1R: dominant black and recessive black alleles segregate in native Swedish sheep populations. Anim. Genet. 50:712–717. doi: 10.1111/age.12837 [DOI] [PubMed] [Google Scholar]
- Shi, X., Wu J., Lang X., Wang C., Bai Y., Riley D. G., Liu L., and Ma X.. . 2021. Comparative transcriptome and histological analyses provide insights into the skin pigmentation in Minxian black fur sheep (Ovis aries). PeerJ. 9:e11122. doi: 10.7717/peerj.11122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, X., Qu G., Wang D., Wang T., Sai W., Chen Y., Yuan L., and Pang Q.. . 2020. Expression and distribution of bone morphogenetic protein 4 and its antagonist Noggin in the skin of Kazakh sheep (Ovis aries) with a white and brown coat color. Acta Histochem. 122:151539. doi: 10.1016/j.acthis.2020.151539 [DOI] [PubMed] [Google Scholar]
- Switonski, M., Mankowska M., and Salamon S.. . 2013. Family of melanocortin receptor (MCR) genes in mammals-mutations, polymorphisms and phenotypic effects. J. Appl. Genet. 54:461–472. doi: 10.1007/s13353-013-0163-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swope, V. B., and Abdel-Malek Z. A.. . 2018. MC1R: front and center in the bright side of dark eumelanin and DNA repair. Int. J. Mol. Sci. 19:2667. doi: 10.3390/ijms19092667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura, K., Peterson D., Peterson N., Stecher G., Nei M., and Kumar S.. . 2011. MEGA5:molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28:2731–2739. doi: 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Våge, D. I, Klungland H., Lu D., and Cone R. D.. . 1999. Molecular and pharmacological characterization of dominant black coat color in sheep. Mamm. Genome. 10:39–43. doi: 10.1007/s003359900939 [DOI] [PubMed] [Google Scholar]
- Weir, B. S., and Cockerham C. C. . 1984. Estimating F-statistics for the analysis of population structure. Evolution. 38:1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x [DOI] [PubMed] [Google Scholar]
- Yang, G. L, Fu D. L, Lang X., Wang Y. T., Cheng S. R., Fang S. L., and Luo Y. Z.. . 2013. Mutations in MC1R gene determine black coat color phenotype in Chinese sheep. Sci. World J. 2013:1675382–1675388. doi: 10.1155/2013/675382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao, L., Bao A., Hong W., Hou C., Zhang Z., Liang X., and Aniwashi J.. . 2019. Transcriptome profiling analysis reveals key genes of different coat color in sheep skin. PeerJ. 7:e8077. doi: 10.7717/peerj.8077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Q., Liu H., and Bu F. X. . 2021. High performance of a GPU-accelerated variant calling tool in genome data analysis. bioRxiv doi: 10.1101/2021.12.12.472266 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.