FIG 1.
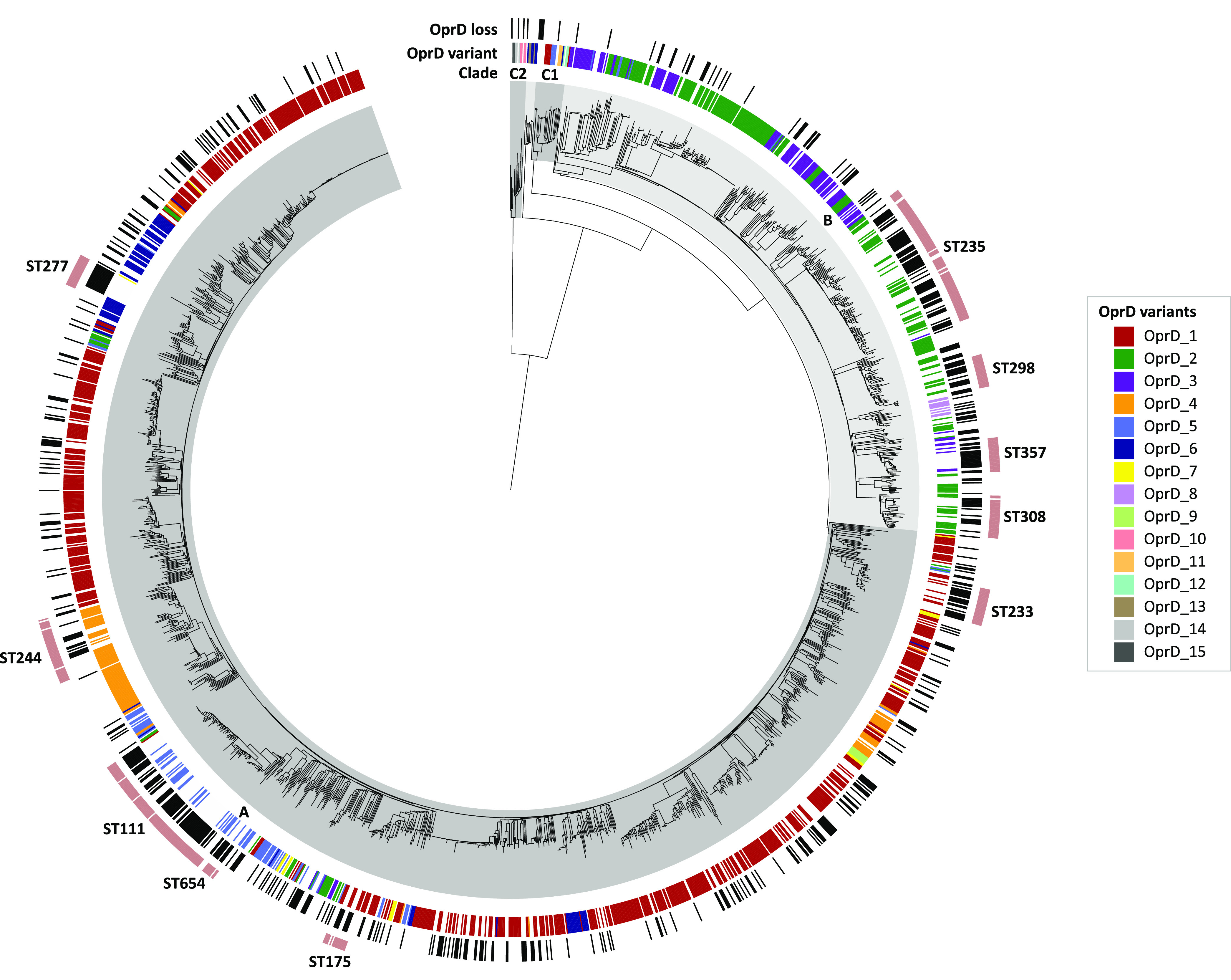
Distribution of OprD variants and predicted OprD loss across the P. aeruginosa phylogeny. Assemblies of 2,088 isolates (NCBI data set) were included in the analysis. Presence of OprD variants (exact amino acid sequence match) is labeled according to the legend. Putative OprD loss is shown in black bars. Intact nonexact matches (n = 51) and putative OprD losses due to promoter disruptions (n = 12) are not indicated. Isolate affiliation to clades (shaded in gray) and high-risk clones (outermost ring) is annotated. Phylogenetic distances were estimated using MASH and visualized in iTOL (46).
