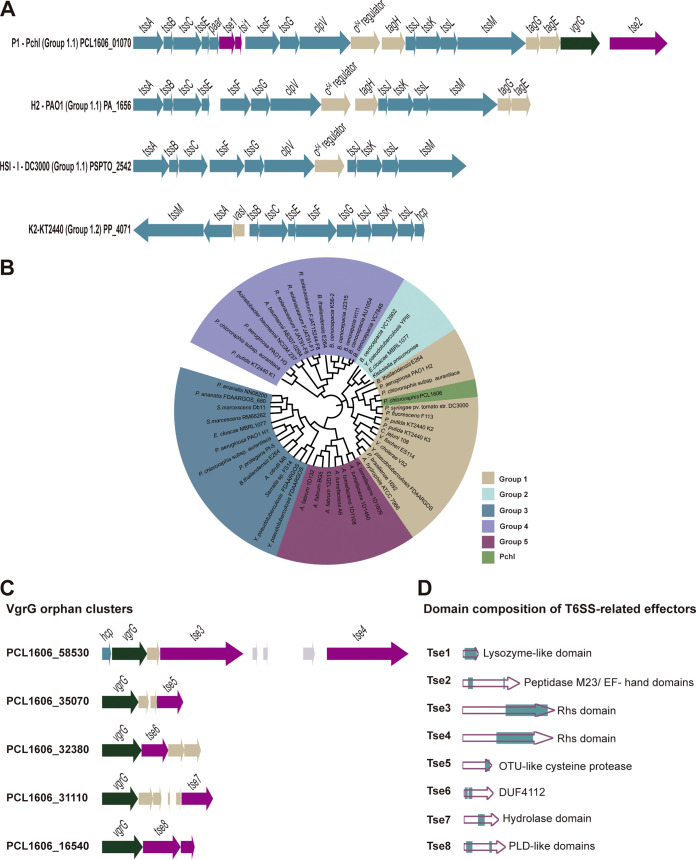
FIG 1.
Characterization of the P. chlororaphis T6SS. (A) Genetic architecture of various Pseudomonas group 1 T6SS clusters. Schematic representations of the main Pchl T6SS cluster, the H2 T6SS cluster of P. aeruginosa PAO1, the HS1-1 T6SS of P. syringae pv. tomato DC3000, and the K2 T6SS of P. putida KT2440. (B) Phylogenetic distribution of T6SS clusters in five main groups based on the tssB locus. Phylogenetic analysis was done using previous nomenclature (107, 108) based on the core protein TssB, widely used as structural component for T6SS phylogenetic analysis (33, 109). The maximum-likelihood method with 1,000 bootstrap replicates was built with Mega X (110) using Clustal Omega (97). The final representation was built using Intel (98). Group 1 is represented in brown, group 2 in light blue, group 3 in dark blue, group 4 in purple, and group 5 in pink. Pchl (green) belongs to group 1. (C) Schematic representation of the VgrG orphan clusters of Pchl. (D) Schematic representation of the predicted domains of eight putative T6SS effectors. Genes encoding structural T6SS proteins are in blue, transcriptional genes in brown, adaptor genes in light brown, VgrG genes in green, and effectors/immunity genes in pink. All arrows representing T6SS genes and the domain sizes of putative effectors are to scale. Full names and accession numbers of strains used to generate T6SS phylogenetic tree in panel B are available in Data Set S1. In addition, Data Set S2 shows the output of a BLASTP analysis comparing the Tse1 amino acid sequence against the NCBI database.