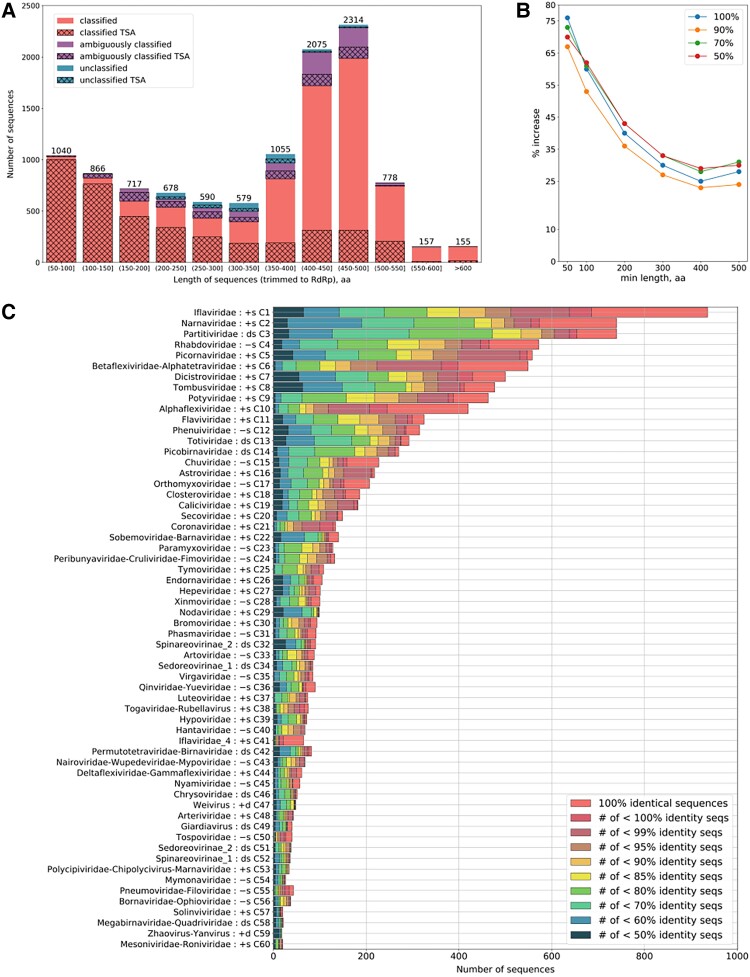
Fig. 1.
Various metrics of identified sequences. (A) Numbers of identified RdRp-encoding ORFs (ref, nr/nt, and TSA) and their lengths after trimming to the RdRp core (see main text) and removing duplicate identical sequences. (B) Percentage increase in the number of RdRp clusters as a function of trimmed RdRp core fragment length (x-axis) and clustering identity threshold, upon adding the TSA-derived sequences to the nr/nt and ref sequences. The y-axis shows the percentage increase in clusters after using different CDHIT (Li and Godzik 2006; Fu et al. 2012) identity thresholds (50%, 70%, 90%, and 100%, as indicated) for nr/nt + TSA sequences compared with nr/nt sequences alone. (C) Numbers of sequences identified in each cluster at different pairwise amino acid identity thresholds. Duplicate identical sequences were removed. Identities were calculated via pairwise alignment in Biopython (Cock et al. 2009, see Materials and Methods) and dividing the number of identical aligned residues by the shorter sequence length.