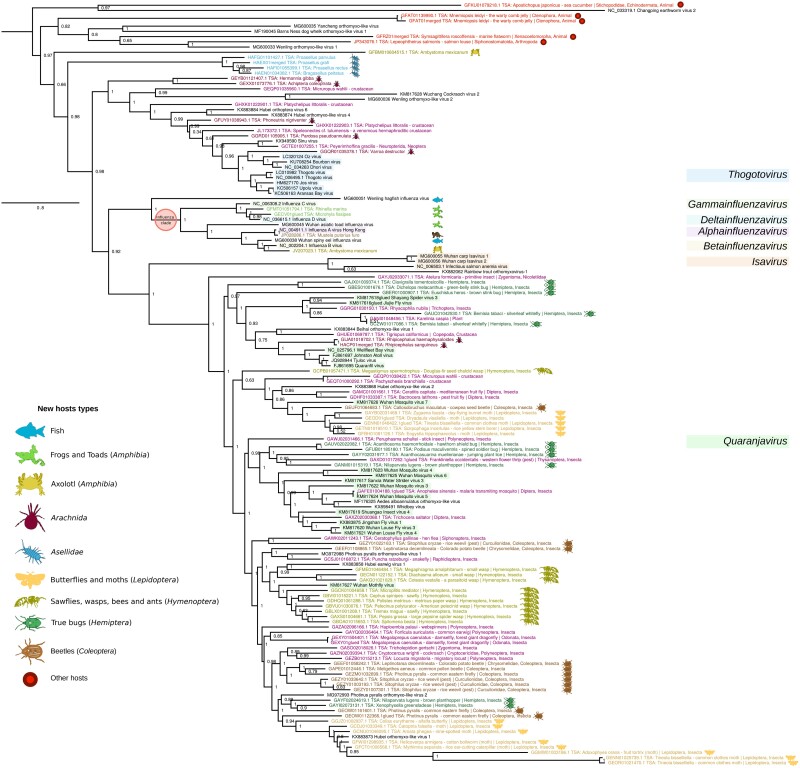
Fig. 7.
Phylogenetic tree of sequences (classified or unclassified) with best match to the orthomyxovirus-like pHMM. Sequences shorter than 100 a.a. were removed, and then, sequences with >95% identity were clustered, and only the longest sequence in each cluster was retained as a representative. NCBI accession numbers, virus names, TSA target organism names, and group-representative icons/colors are shown (key at left). Currently defined genera are identified with colored highlighting (key at right). Sequences labeled as “merged” derive from merged overlapping contigs. Sequences labeled as “glued” comprise multiple concatenated ORFs from a contig that was inferred to likely have sequence quality issues which introduce stop codons (e.g., via frameshift errors—common with 454 sequencing) or potentially derive from mutated endogenized viral elements, EVEs). Ferret icon has been added but it is a well-known host type.