FIG 1.
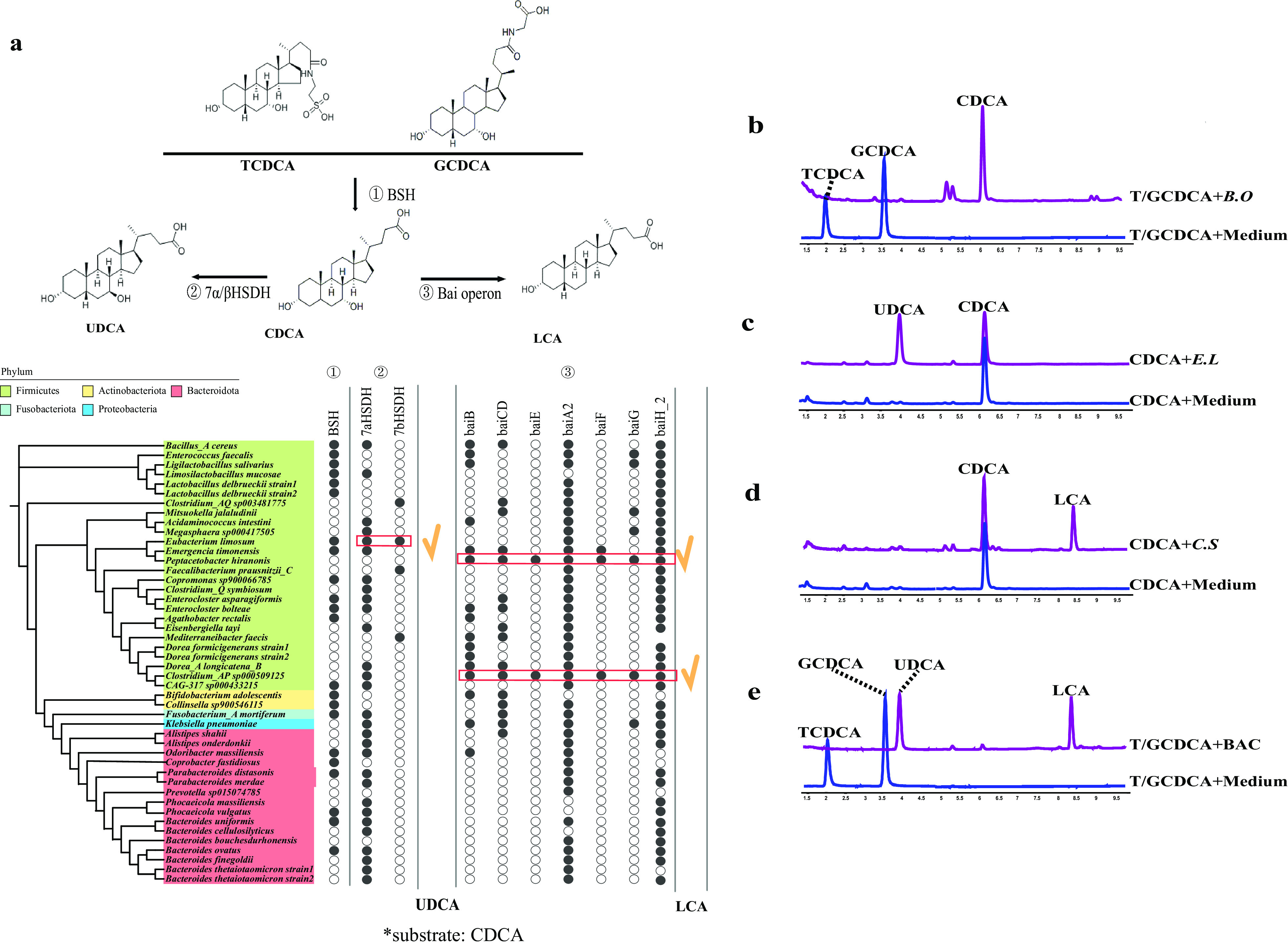
Bioinformatics analysis and experimental validation of bile acid metabolism of human gut bacterial strains. (a) Homology-based search in the whole-genome sequence of human gut bacterial strains identified genes related to bile acid metabolism, including BSH, 7α/βHSDH, and the bai operon. Red rectangles, E. limosum with 7α/βHSDH, Clostridium AP sp000509125, and Peptacetobacter with the complete bai operon. (b) B. ovatus converts TCDCA and GCDCA into CDCA. (c) E. limosum converts CDCA into UDCA. (d) Clostridium AP sp000509125 converts CDCA into LCA. (e) The three-strain consortium BAC (B. ovatus, E. limosum, and Clostridium AP sp000509125) converts TCDCA and GCDCA into UDCA and LCA. In our experimental validation, bacterial strains were incubated with TCDCA and GCDCA or CDCA for 60 h and then subjected to LC/MS description. G, glyco-; T, tauro-; UDCA, ursodeoxycholic acid; LCA, lithocholic acid; CDCA, chenodeoxycholic acid; B.O, Bacteroides ovatus; E.L, Eubacterium limosum; C.S, Clostridium AP sp000509125.
