Fig. 2 |. Comparison of unliganded, cargo-bound and RAN-bound importins.
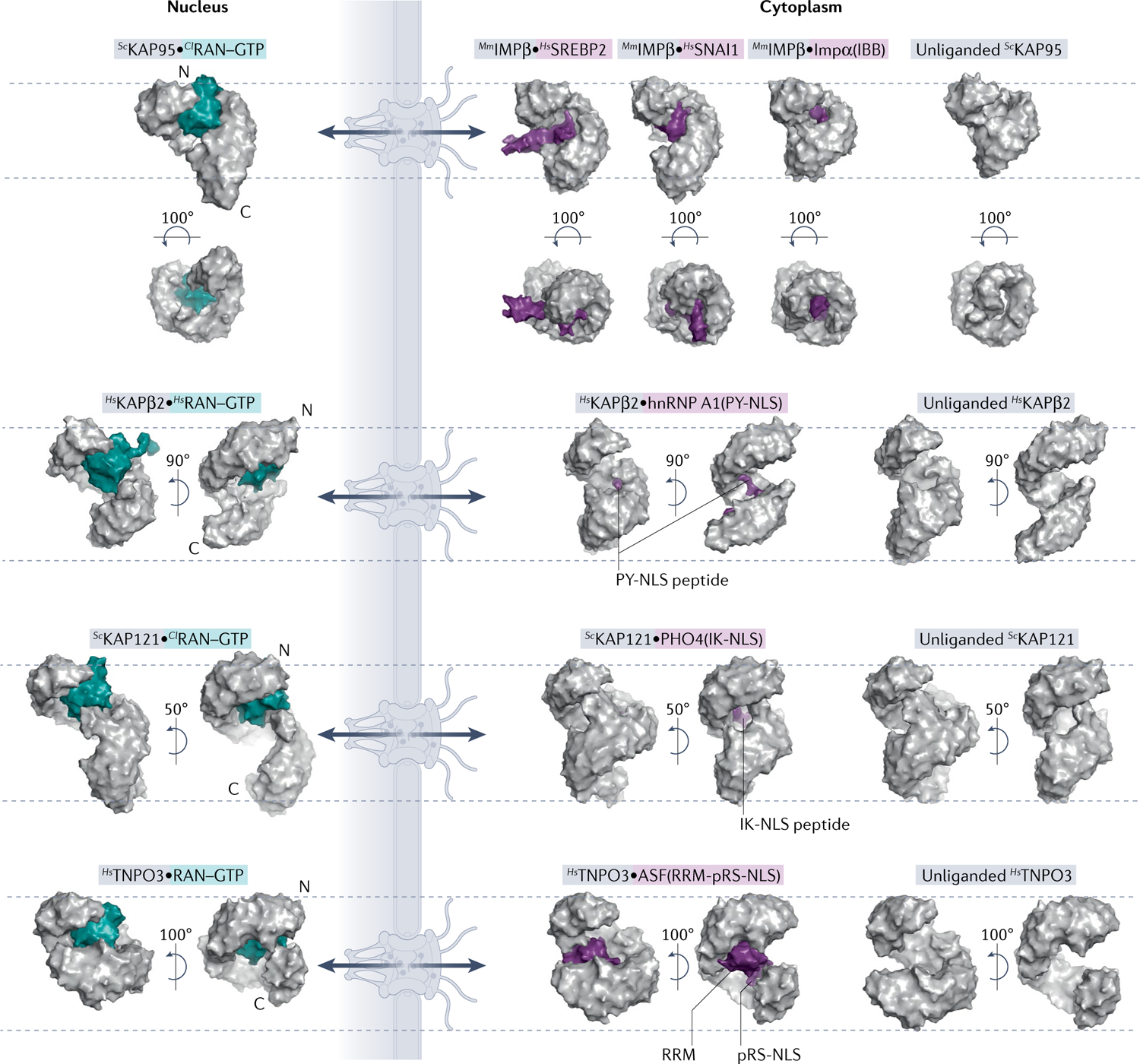
Surface representations of nuclear and cytoplasmic complexes of importins (grey) with RAN–GTP (teal) or cargo (purple). All importins are structurally aligned at residues 1–200 and displayed in the same orientation. Dashed lines mark top of first HEAT repeat and bottom of last HEAT repeat of each unliganded importin to help visualize importin conformational changes and differences in superhelical compactness. Cargo release by RAN–GTP results from either steric clash between RAN–GTP and cargo or conformational changes to the importin superhelix. PDB IDs (left to right): IMPβ [PDB:2BKU, PDB:1UKL, PDB:3W5K, PDB:1QGK, PDB:3ND2]; KAPβ2 [PDB:1QBK, PDB:2H4M, PDB:2QMR]; KAP121 [PDB:3W3Z, PDB:3W3X, PDB:3S3T]; and TNPO3 [PDB:4C0Q, PDB:4C0O, PDB:4C0P]. ASF, splicing factor 1; Cl, Canis lupus; hnRNP, heterogeneous nuclear RNP; Hs, Homo sapien; IBB, amino-terminal IMPβ-binding; IK-NLS, isoleucine-lysine nuclear localization signal; Kaps, Karyopherins; Mm, Mus musculus; PY-NLS, proline-tyrosine nuclear localization signal; RRM, RNA-recognition motif; RS-NLS, arginine-serine repeat nuclear localization signal; Sc, Saccharomyces cerevisiae; SREBP2, sterol regulatory element-binding protein 2.
