Extended Data Fig. 7 |. ETV6 antagonizes EWS-FLI1 function at select DNA elements.
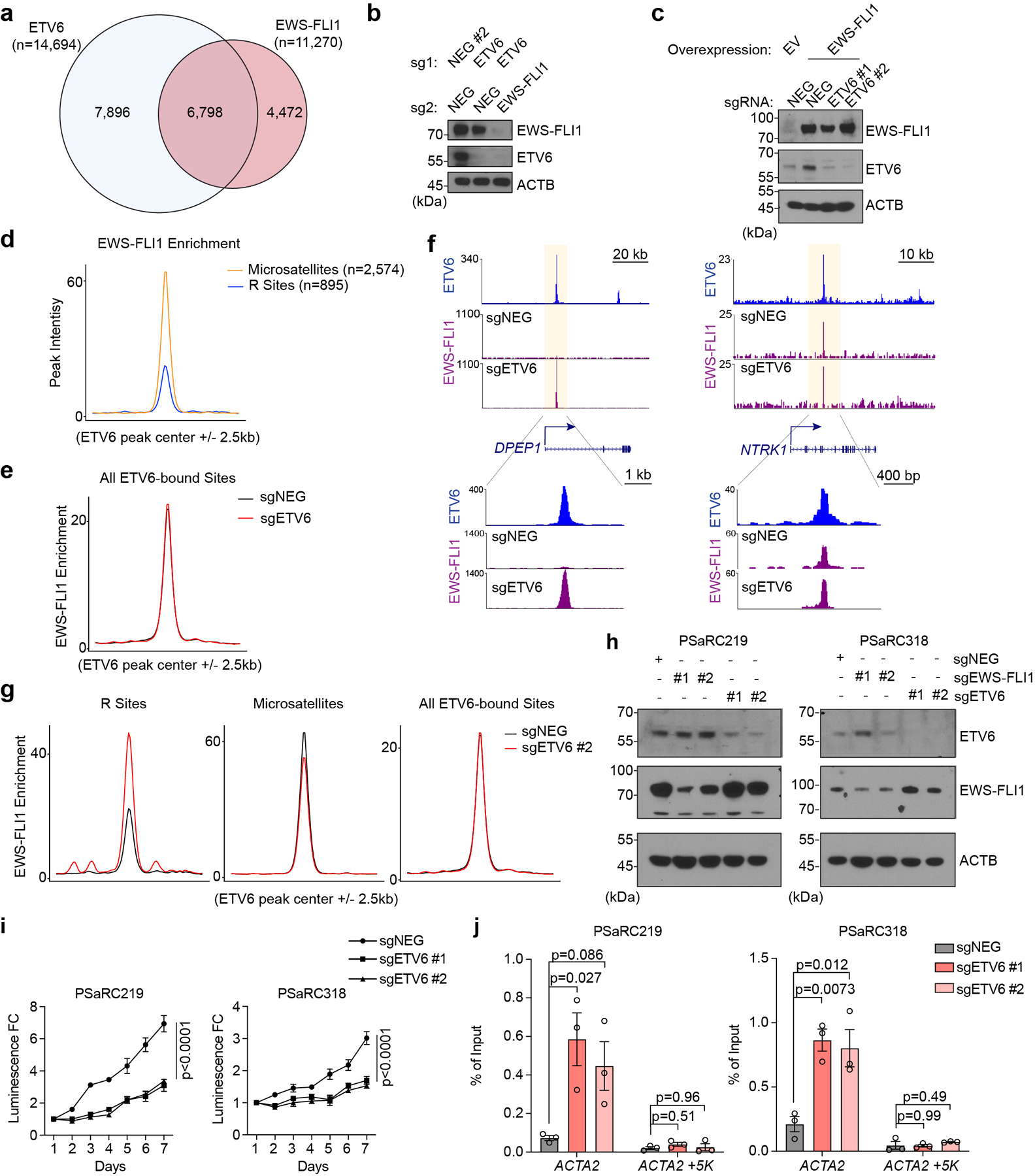
(a) Venn diagram showing the overlap between ETV6 and EWS-FLI1 peaks in A673 cells. (b)Western blot assessing EWS-FLI1 and ETV6 levels in A673 cells transduced with indicated dual-sgRNA constructs. (c)Western blot of EWSFLI1 and ETV6 in EWS-FLI1 reprogrammed RD cells. RD cells were transduced with EV (empty vector) control or EWS-FLI1 linked to indicated sgRNAs. (d) Metagene representation of the mean EWS-FLI1 signal differences at R sites versus microsatellites. (e) Metagene representation of the mean EWS-FLI1 signal differences at all ETV6 binding sites in A673 cells upon targeting ETV6. (f) Gene tracks of ETV6 and EWS-FLI1 ChIP-seq occupancy upon ETV6 knockout at DPEP1 and NTRK1 sites at two different scales. (g) Metagene representation of mean EWS-FLI1 ChIP-seq signal changes across R sites, microsatellites and all ETV6 binding regions in A673 cells upon targeting ETV6 using sgETV6 #2. (h) Western blot of ETV6 and EWS-FLI1 levels in patient-derived tumour cells (PSaRC219, left; PSaRC318, right) infected with indicated sgRNAs. ACTB was used as a loading control. (i) CellTiter Glo assay evaluating the effects of ETV6 knockout to proliferation in the patient-derived tumour cells (PSaRC219, left; PSaRC318, right). The relative luminescence fold changes (FC) to day 1 were plotted. Data are mean ± SEM. P values were calculated using two-way ANOVA. (n = 4 biological replicates) (j) EWS-FLI1 ChIP-qPCR analysis assessing the EWS-FLI1 occupancy changes at the ACTA2 site in the patient-derived Ewing tumour cells (PSaRC219 and PSaRC318) upon ETV6 knockout. A site 5 kb downstream of the ACTA2 site was used as a negative control. Data are mean ± SEM. P values were calculated using one-way ANOVA, Dunnett’s multiple comparison tests. (n = 3 biological replicates).
