Figure 1. The pathogen-derived metabolite phenazine-1-carboxamide (PCN) activates anti-pathogen defenses in the C. elegans intestine.
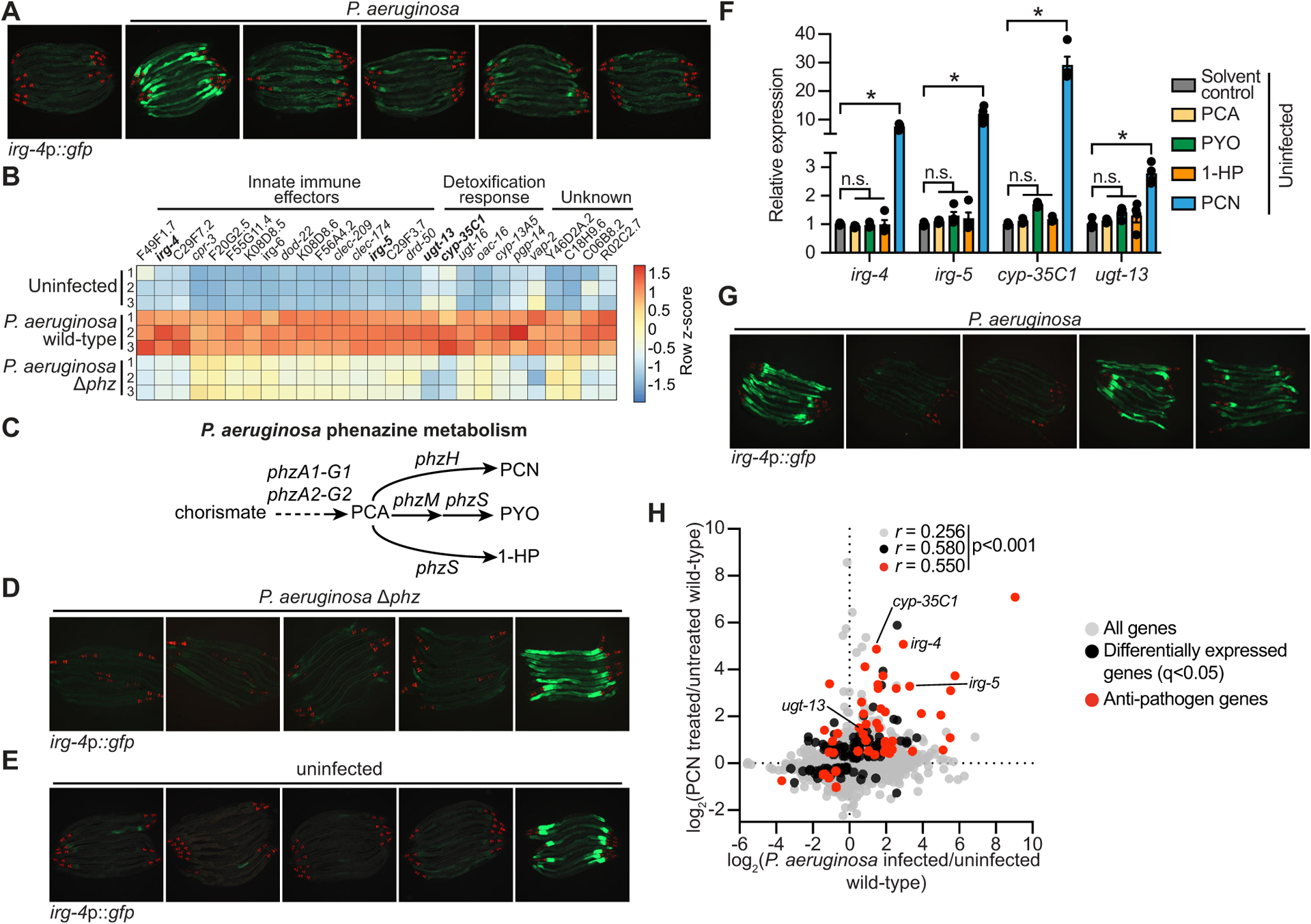
(A) Images of C. elegans irg-4p∷gfp transcriptional reporter expression in animals either uninfected or infected with the indicated P. aeruginosa strains, (scale bar, 200 μM). (B) Heat map of the 27 genes that are induced in C. elegans during P. aeruginosa infection in a manner dependent on the production of phenazines (q<0.05). Gene expression from biological replicates in each condition were scaled by calculating the row z-score for each gene (n=3). See also Table S1A. (C) A schematic of P. aeruginosa phenazine metabolism (PCA, phenazine-1-carboxylic acid; PCN, phenazine-1-carboxamide; PYO, pyocyanin; 1-HP, 1-hydroxyphenazine). (D and E) Images of C. elegans irg-4p∷gfp animals during infection with P. aeruginosa Δphz (D) or grown under standard conditions (uninfected) (E) on media that was supplemented with the indicated phenazines, (scale bar, 200 μM). (F) qRT-PCR analysis of the indicated anti-pathogen genes in wild-type animals exposed to the indicated phenazines in the absence of infection. Data are the average of biological replicates with error bars giving SEM (n=4). *equals p<0.05 (Brown-Forsythe and Welch ANOVA with Dunnett’s T3 multiple comparisons test). Concentration of phenazines used in (D), (E) and (F) are 112 μM PCN, 112 μM PCA, 119 μM PYO, and 25 μM 1-HP. (G) Images of C. elegans irg-4p∷gfp animals infected with the indicated P. aeruginosa strains. (scale bar, 200 μM). (H) Data from mRNA-sequencing experiments comparing genes differentially regulated in wild-type animals exposed to PCN (y-axis) with genes differentially expressed in wild-type animals during P. aeruginosa infection (x-axis). All genes are shown in gray. Genes that are differentially expressed in both datasets are shown in black (q<0.05), and the differentially expressed genes annotated as anti-pathogen genes (innate immune effector or detoxification genes) are shown in red. The Pearson correlation coefficient (r) between the indicated transcriptional signatures is shown. The location of the genes irg-4, irg-5, cyp-35C1, and ugt-13, whose regulation is examined throughout this manuscript, are shown. See also Table S1B. Source data for this figure is in Table S3. See also Fig. S1.
