Figure 5. The bacterial metabolite PCN is a pattern of pathogenesis sensed by C. elegans NHR-86 to activate innate immunity.
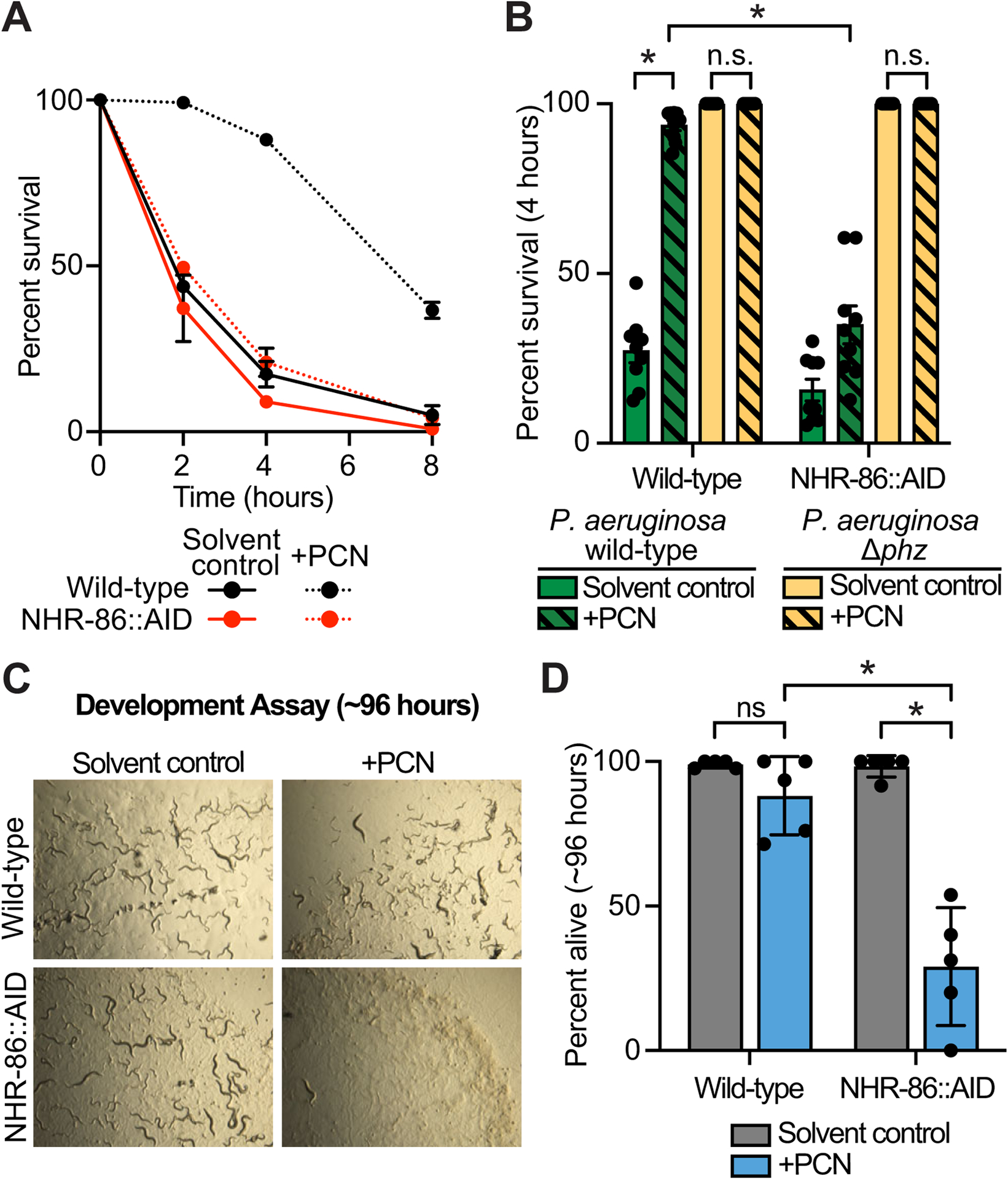
(A) A phenazine toxicity assay in C. elegans (also called the “fast kill” assay) with P. aeruginosa and C. elegans of the indicted genotypes either treated with solvent control or PCN (448 μM). Data are representative of three trials. The difference between PCN-treated wild-type and NHR-86∷AID animals is significant (p<0.05, log-rank test, n=3). Survival curves for these strains exposed to the P. aeruginosa Δphz mutant are shown in Fig. S5A. Sample sizes, four-hour survival, and p-values for each replicate are shown in Table S2. (B) Survival data at four hours after exposure to the indicated conditions is shown for the experiment described in (A). Data are the average of three biological replicates each containing three trials with error bars showing SEM (n=9). *equals p<0.05 (two-way ANOVA with Tukey’s multiple comparisons test). Sample sizes, four-hour survival, and p-values for each replicate are shown in Table S2. (C and D) A development assay with wild-type and NHR-86∷AID C. elegans. Animals of the indicated genotypes were allowed to lay their brood in the presence or absence of PCN, as indicated, and (C) photographed after approximately 96 hours or (D) scored for the number of alive animals (n=5). All assay plates contained 50 μM auxin. *p<0.05 for the indicated comparisons (two-way ANOVA with Šídák’s multiple comparisons test). n.s.=not significant. Source data for this figure is in Table S3. See also Fig. S5.
