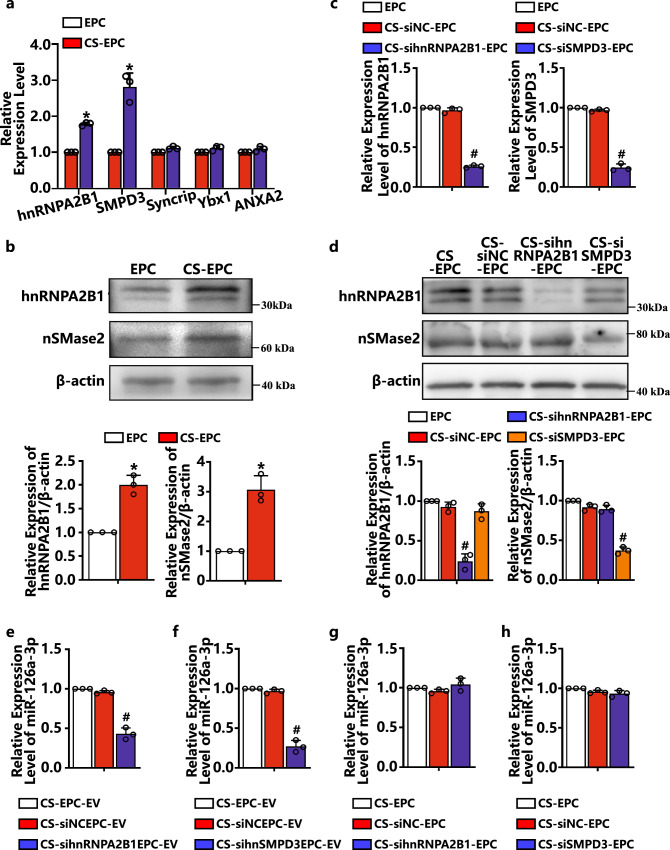
Fig. 9. miR-126a-3p was mediated by Heterogeneous Nuclear Ribonucleoprotein A2/B1 (hnRNPA2B1) and Neutral Sphingomyelinase 2 (nSMase2) to sort into EVs (n = 3, biological replicates per group).
a The expression of hnRNPA2B1, Sphingomyelin Phosphodiesterase 3 (SMPD3), Syncrip, Y-box binding protein 1 (Ybx1), and Annexin A2 (ANXA2) in EPCs after culture with silicate ions (CS-EPCs) for 48 h. b Western blot analysis of hnRNPA2B1 and nSMase2 proteins in EPCs after culture with silicate ions for 48 h. *p < 0.05 vs. EPC. c The expression of hnRNPA2B1 and SMPD3 in CS-EPCs after knockdown with specific siRNA. d Protein analysis of hnRNPA2B1 and nSNase2 in CS-EPCs after knockdown with specific siRNA. e, f The expression of miR-126a-3p in EVs after knockdown of hnRNPA2B1 (e) and SMPD3 (f). g, h The expression of miR-126a-3p in EPCs after knockdown of hnRNPA2B1 (g) and SMPD3 (h). #p < 0.05 vs. the other two groups. Data are presented as the mean ± standard. Two-tailed Student’s t-test was used to compare the differences between two groups. One-way ANOVA and post hoc Bonferroni tests were used to compare differences among more than two groups. Source data are provided as a Source Data file. Each experiment was repeated 3 times or more independently with similar results.