Abstract
Purpose
To investigate the mechanisms of Klebsiella pneumoniae-induced pyogenic liver abscess (PLA).
Methods
Forty-three K. pneumoniae strains from PLAs and 436 from non-PLAs were collected. Their differences were compared for virulence genes and factors, sequence types, and serotypes. Virulence genes wzi, wzy-K1, and wzi+wzy-K1 were deleted in K. pneumoniae NTUH-K2044. Various analyses, such as transmission electron microscopy, neutrophil killing tests, and mouse lethality tests, were used to confirm the consequent changes.
Results
Differences were found between K. pneumoniae strains from PLA and non-PLA samples for virulence genes and factors, including metabolism genes (allS and peg-344), capsular polysaccharide (CPS)-synthesis channel gene (wzy-K1), CPS-regulating genes (p-rmpA, p-rmpA2, and c-rmpA), and siderophore genes (iucA and iroN). When wzy-K1 was positive, the difference between PLA and non-PLA samples was only observed with c-rmpA. Δwzi, Δwzy-K1, and ΔwziΔwzy-K1 strains reverted to hypovirulence. In the Kupffer cell stimulation assay, interleukin (IL)-6, IL-12, IL-10, and transforming growth factor-β secretions were found to be equivalent in NTUH-K2044, Δwzi, Δwzy-K1, and ΔwziΔwzy-K1 groups. Lower IL-1β and higher tumor necrosis factor-α secretions were observed for Δwzi, Δwzy-K1, and ΔwziΔwzy-K1.
Conclusions
Hypercapsule production is the cornerstone of hypervirulence, regardless of exopolysaccharides. K1 K. pneumoniae-induced PLA may decrease core inflammatory cytokines rather than increase anti-inflammatory cytokines. Exopolysaccharides could also attenuate the inflammatory response to aid in the immune escape of K. pneumoniae.
Keywords: Klebsiella pneumoniae, pyogenic liver abscess, mouse lethality test, hypervirulence, mechanism
Introduction
Pyogenic liver abscess (PLA) is a common infectious disease in clinical practice (Meddings et al., 2010). PLA was first diagnosed and presented by Ochsner et al. in 1938 and is known to be induced by many kinds of pathogens (Roediger and Lisker-Melman, 2020). The morbidity rate of PLA increased from 11/100,000 to 18/100,000 in Taiwan between 1998 and 2004 (Tsai et al., 2008). In Singapore and Mainland China, 86 – 160 patients suffer from PLA per 100,000 hospitalized patients (Tian et al., 2012; Lo et al., 2015). The mortality rate accounts for 13.0% in patients with PLA (Lo et al., 2015). Therefore, PLA is a fatal disease, whose incidence is increasing and becoming a concern.
Bacteria account for the majority of PLA cases, 85.1% of which are caused by Klebsiella pneumoniae and Escherichia coli strains with a ratio of almost 6: 1 (Tian et al., 2012; Shi et al., 2017). K. pneumoniae exists widely in nature, including healthcare settings (Podschun et al., 2001). In humans, K. pneumoniae can colonise various sites, such as the axilla, intestines, and nasopharynx (Lin et al., 2012). K. pneumoniae can induce nosocomial infections, such as pneumonia, sepsis, and urinary tract infections; it can also cause community-acquired infections, such as PLA, necrotising fasciitis, endophthalmitis, and meningitis (Russo and Marr, 2019). K. pneumoniae can be classified as hypervirulent K. pneumoniae (HvKP) and classical K. pneumoniae (cKP) (Russo and Marr, 2019). HvKP often possesses extreme resistance to serum and human neutrophils (Gu et al., 2018). K. pneumoniae strains causing PLA are mostly hypervirulent, particularly those typed as K1 and K2 (Qu et al., 2015). However, to our knowledge, the mechanisms of development of K. pneumoniae-induced PLA are currently unknown.
K. pneumoniae can harbour multiple virulence factors, such as exopolysaccharide (EPS), capsular polysaccharide (CPS), lipopolysaccharide, siderophores, fimbriae, allantoin metabolism, outer membrane proteins, and porins (Paczosa and Mecsas, 2016). These factors contribute to the virulence of K. pneumoniae, such as the anti-phagocytosis, anti-complement, and anti-biofilm effects of CPS, and are encoded by a series of virulence genes. For instance, CPS synthesis is controlled by the cps cluster and is regulated by the regulator of mucoid phenotype A gene (rmpA). In addition, CPS anchoring is dependent on the gene wzi (Paczosa and Mecsas, 2016). Therefore, K. pneumoniae virulence has considerable complexity, which needs to be studied further.
The purpose of the present study was to analyse important virulence factors in K. pneumoniae by comparing gene positivity in PLA-inducing and non-PLA-inducing strains and by determining the effect of deletions of specific genes on K. pneumoniae virulence, focusing on genes related to CPS and EPS.
Materials and methods
K. pneumoniae strains
In this study, 43 and 436 K. pneumoniae strains were obtained from PLA and non-PLA patients, respectively. All the strains were isolated from distinct patients. The PLA strains were isolated from abscess, drainage, and puncture fluid specimens at the Department of Infectious Diseases, the First Affiliated Hospital of Zhejiang University in 2017. PLA was diagnosed based on pathogen and imaging evidences (B-mode ultrasonography and computed X-ray tomography). The non-PLA strains were obtained from seven hospitals in China between January 2017 and February 2018: Huashan Hospital, 180 strains; Jinshan Hospital, 28 strains; Taizhou Municipal Hospital, 84 strains; the First Affiliated Hospital of Guangxi Medical University, 20 strains; Kunming Yan’an Hospital, 34 strains; Sixth Hospital of Shanxi Medical University, 60 strains; Shandong Provincial Hospital Affiliated with Shandong University, 30 strains. Their sources included sputum (225, 58.5%), urine (98, 22.5%), blood (29, 6.7%), and others (54, 12.4%).
The specimens were stored at -80°C until use. Sheep blood agar plates were used to culture the strains, followed by identification using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (Bruker Daltonics Inc., Fremont, CA, USA). The standard strains Pseudomonas aeruginosa ATCC 27853, K. pneumoniae ATCC 700603, and E. coli ATCC 25922 were used as controls.
K. pneumoniae NTUH-K2044 (Accession number: AP006725.1), a typical HvKP, was originally isolated from the Department of Internal Medicine, National Taiwan University Hospital, Taipei, Taiwan. K. pneumoniae HS11286 (Accession number: CP003200.1), a K47 strain expressing bla KPC and showing hypovirulence, was originally isolated from the Department of Laboratory Medicine, Huashan Hospital, Fudan University, Shanghai, China. Strains NTUH-K2044 and HS11286 were used as controls for strain morphological tests, string tests, capsule staining, periodic acid–Schiff staining, fitness analyses, quantitative PCR, mouse lethality tests, and Galleria mellonella lethality tests. Strain NTUH-K2044 was also a target for gene deletions (wzi, wzy-K1, and wzi+wzy-K1). The genetic traits of NTUH-K2044 and HS11286 were shown in Table 1 .
Table 1.
Genetic backgrounds of NTUH-K2044 and HS11286.
| Strain | ST | Serotype | allS | peg-344 | wzy-K1 | entB | irp2 | iroN | iucA | fimH | mrkD | c-rmpA | p-rmpA | p-rmpA2 | wzi |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NTUH-K2044 | 23 | K1 | + | + | + | + | + | + | + | + | + | + | + | + | + |
| HS11286 | 11 | K47 | – | – | – | + | + | – | – | + | + | – | – | – | + |
ST, sequence type; +, positive; -, negative.
The study was a retrospective investigation; thus, approval to use the 43 and 436 K. pneumoniae strains was waived.
Multilocus sequence typing
The primers of seven housekeeping genes (gapA, infB, mdh, pgi, phoE, rpoB, and tonB) are shown in Table S1 . The QIAamp DNA mini kit (catalogue number: 51304, QIAGEN, Düsseldorf, Germany) was used to extract DNA from the K. pneumoniae strains following the manufacturer’s protocol. PCR was used to amplify the housekeeping genes, and the products were sequenced using an ABI 3730XL DNA Analyzer (Applied Biosystems, San Ramon, CA, USA). Sequence types (STs) were obtained after searching the K. pneumoniae MLST database (http://bigsdb.pasteur.fr/cgi-bin/bigsdb/bigsdb.pl?db=pubmlst_klebsiella_seqdef&page=sequenceQuery).
Determination of serotypes and virulence genes
The wzi locus was sequenced to determine serotypes of the K. pneumoniae strains by searching the Institute Pasteur database (http://bigsdb.pasteur.fr/cgi-bin/bigsdb/bigsdb.pl?db=pubmlst_klebsiella_seqdef&page=sequenceQuery).
Virulence genes, wzy-K1, allS, entB, irp2, iroN, iucA, fimH, mrkD, p-rmpA, p-rmpA2, c-rmpA, peg-344, and wzi (Compain et al., 2014; Gu et al., 2018; Russo et al., 2018), were analysed using a Veriti PCR system (Applied Biosystems). The primers used are shown in Table S1 . NTUH-K2044 was used as the positive control. In the subsequent agarose electrophoresis, a proper band in line with the control is regarded as a positive gene.
Gene deletions in K. pneumoniae NTUH-K2044
Deletions of wzi+wzy-K1, and wzi+wzy-K1 were constructed using the lambda Red recombination method as previously described (Datsenko and Wanner, 2000). The primers used are shown in Table S1 .
String test
String tests were performed thrice per strain as previously described (Shon et al., 2013). The test was considered positive when the produced string was longer than five mm.
Capsule staining
K. pneumoniae strains were stained to distinguish their hypercapsules according to the manufacturer’s instructions (catalogue number: BA-4039; BASO, Zhuhai, China).
Periodic acid–Schiff staining
Periodic acid–Schiff staining was performed to detect EPS according to the manufacturer’s protocol (catalogue number: BA4080A; BASO, Zhuhai, China).
Transmission electron microscopy
TEM was performed using a Tecnai G2 Spirit Twin Electron Microscope (FEI, Hillsboro, USA), as previously reported (Ernst et al., 2020). K. pneumoniae strains were cultured overnight on sheep blood agar plates. Luria Bertani (LB) broth was then used to culture them to mid-log phase. The strains were collected after centrifugation at 11000 g for 5 min. The sediments were immersed at 2.5% glutaraldehyde overnight, followed by washing thrice with phosphate buffered saline (PBS) and fixation with 1% osmic acid for 1.5 h. The pellets were then washed thrice with PBS, followed by dehydration with grades of alcohol (30%, 50%, 70%, 80%, 95%: 15 min; 100%: 2 × 10 min). Infiltration and embedding were performed using acetone and Epon-812 respectively, followed by polymerization at 60°C for 48 h. The pellets were sliced and stained by 2% uranyl acetate water (10 min) and lead citrate (10 min) before TEM.
Human neutrophil killing assay
This protocol is based on a previously reported method (Deleo et al., 2014). The human neutrophil killing assay was approved by the Ethics Committee of Huashan Hospital (Shanghai, China). Neutrophils (1 × 106) from healthy volunteers and 1 × 106 colony forming unit (CFU) of opsonized K. pneumoniae were mixed in RPMI/H medium at 37°C for zero and 60 min with gentle rotation. One percent saponin was added to each tube, mixed, and chilled on ice for 15 min. The mixture was diluted and cultured overnight on LB agar plates. Viable colonies were counted, and the survival ratios were calculated relative to the zero min point. Each strain was tested three times.
Serum killing assay
This protocol is based on a previously reported method (Lin et al., 2014). The serum killing assay was approved by the Ethics Committee of Huashan Hospital (Shanghai, China). Human blood was collected from ten healthy volunteers, and sera were isolated. The sera were mixed, divided into 500 μL aliquots, and stored at -80°C prior to use. Mid-log-phase K. pneumoniae strains were washed twice with normal saline and readjusted to 4 × 106 CFU/mL, 25 μL of which were added to 75 μL of pooled sera in a 12-well plate (Corning Incorporated, Corning, NY). After zero, one, two, and three h of incubation at 37°C, the mixture was diluted and cultured overnight on LB agar plates. Viable colonies were counted, and the survival ratio was calculated relative to the zero h point. Each strain was tested three times.
Fitness analysis
Fitness of the K. pneumoniae strains was determined as previously described (Liu et al., 2016). Overnight cultures were diluted to an optical density at 600 nm (OD600) of 0.001 and incubated at 37°C under aerobic conditions, monitoring the OD600 every 30 min on a microplate reader (BioTek Synergy H1, Winooski, VT, USA).
Real-time quantitative PCR
Real-time quantitative PCR on an Applied Biosystems 7500 System was used to evaluate the expression of p-rmpA, p-rmpA2, c-rmpA, manC, and galF, with 16S rRNA as the reference gene. The primers used are shown in Table S1 . The analyses were performed according to the manufacturer’s protocol of the SYBR Green qPCR Mix (catalogue number: FS-Q1002; FOREVER STAR, Beijing, China).
Mouse lethality test
Six-week-old pathogen-free female BALB/c mice (four per group) were inoculated intraperitoneally with 100 μL of the K. pneumoniae strains (102–107 CFU, mid-logarithmic growth) that had been washed twice with normal saline (Mizuta et al., 1983). The mice were then observed for 14 days. Lethality dose 50 (LD50) values and survival curves were obtained as reported in a previous study (Reed and Muench, 1938).
Kupffer cell stimulation assay
The Kupffer cells were isolated from specific pathogen-free mice and the cell stimulation assays were performed as described previously (Lalitha et al., 2017; Hoh et al., 2019). Six hundred microlitres of 1 × 104 Kupffer cells were inoculated into each well of a 48-well plate, followed by the addition of 2.0 μL of lipopolysaccharide (1.5 mg/mL) and interferon (0.25 mg/mL). After incubation at 37°C in a 5.0% CO2 atmosphere for two h, 20 μL of a K. pneumoniae strain (5 × 106 CFU/mL) were added, resulting in a multiplicity of infection of 10. After four, eight, and twelve h of further incubation, the suspensions were collected and centrifuged at 10621g for ten min. Interleukin (IL)-1β, IL-6, IL-10, IL-12, tumor necrosis factor-α (TNF-α), and transforming growth factor-β (TGF-β) were measured in the supernatants as indicated in ELISAs.
ELISA
ELISAs were used to detect IL-1β, IL-6, IL-10, IL-12, TNF-α, and TGF-β, according to the manufacturer’s instructions (catalogue numbers: F10770, F10830, F10870, F10880, F11630, and F11591; Shanghai Westang Biotech Inc., Shanghai, China).
Statistical analysis
GraphPad Prism 8 software (GraphPad Software Inc., San Diego, CA, USA) was used for Chi-square tests, Fisher’s exact tests, one-way ANOVA, factorial design analysis of variance, and Kruskal–Wallis tests between groups. Significance was considered at p < 0.05.
Results
Significant differences between K. pneumoniae from PLA and non-PLA
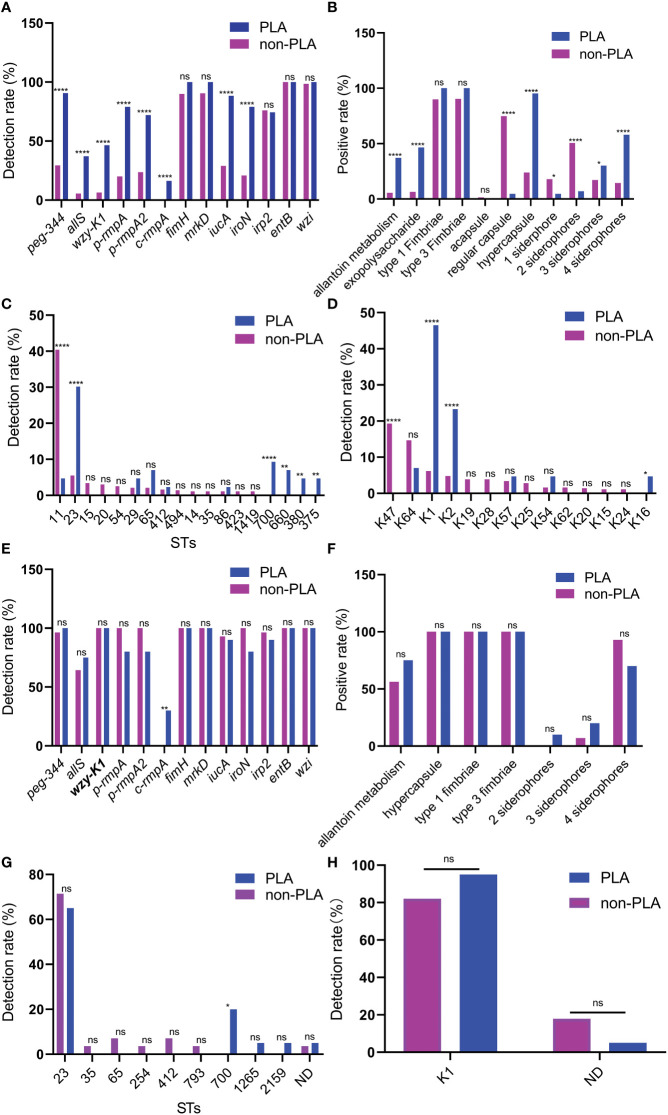
As shown in ( Figures 1A, B ), higher rates were found in K. pneumoniae strains from PLA than those from non-PLA for virulence genes and factors, including metabolism (allS and peg-344), CPS-synthesis channel (wzy-K1), CPS-regulating (p-rmpA, p-rmpA2, and c-rmpA), and siderophore (iucA and iroN) genes. For STs, a lower rate of ST11 and higher rates of ST23/ST700/ST660/ST380/ST375 were observed in strains from PLA; for serotypes, a lower rate of K47 and higher rates of K1/K2/K16 were found in strains from PLA ( Figures 1C, D ).
Figure 1.
Differences between Klebsiella pneumoniae from PLA and non-PLA samples. (A) Differences in virulence genes; (B) Differences in virulence factors; (C) Differences in STs; (D) Differences in serotypes; (E) Differences in virulence genes when wzy-K1 is positive; (F) Differences in virulence factors when wzy-K1 is positive; (G) Differences in STs when wzy-K1 is positive; (H) Differences in serotypes when wzy-K1 is positive. PLA: pyogenic liver abscess; PLA group: n = 43 (A–D) n = 20 (E–H) non-PLA group: n = 436 (A–D) n = 28 (E–H) ns: not significant; *p < 0.05; **p < 0.01; ****p < 0.0001; ST, sequence type; ND, not defined. Chi-square tests and Fisher’s exact tests were used to compare the positive rates between PLA and non-PLA groups.
Since virulence gene wzy-K1 was with significant difference (p < 0.0001) and nearly half-positive (20/43) in strains from PLA, it was used for stratification analysis. When wzy-K1 was positive, significant differences were observed only with c-rmpA (p < 0.01) and ST700 (p < 0.05) ( Figures 1E, G ): higher rates of c-rmpA (6/20) and ST700 (4/20) in strains from PLA; no significant differences were found for virulence factors and serotypes ( Figures 1F, H ). When wzy-K1 was positive, the positivity of virulence genes increased, except for fimH, mrkD, irp2, entB, and wzi ( Figures 1A, E ).
Roles of CPS and EPS in PLA
To investigate the roles of CPS and EPS in PLA, wzi, wzy-K1, and wzi+wzy-K1 were deleted in NTUH-K2044. The colonies were larger with Δwzi and smaller with Δwzy-K1 and ΔwziΔwzy-K1 than with NTUH-K2044 ( Figures 2A–E ). The string test was positive for Δwzi but negative for Δwzy-K1 and ΔwziΔwzy-K1. Capsule staining showed that Δwzi, Δwzy-K1, and ΔwziΔwzy-K1 did not have hypercapsules ( Figures 2F–J ). TEM suggested that Δwzi and ΔwziΔwzy-K1 were without a capsule, while Δwzy-K1 had a thin capsule ( Figures 2K–O ). Periodic acid–Schiff staining confirmed the presence of EPS in Δwzi and NTUH-K2044 and its abscence in Δwzy-K1, ΔwziΔwzy-K1, and HS11286 ( Figures 2P–T ).
Figure 2.
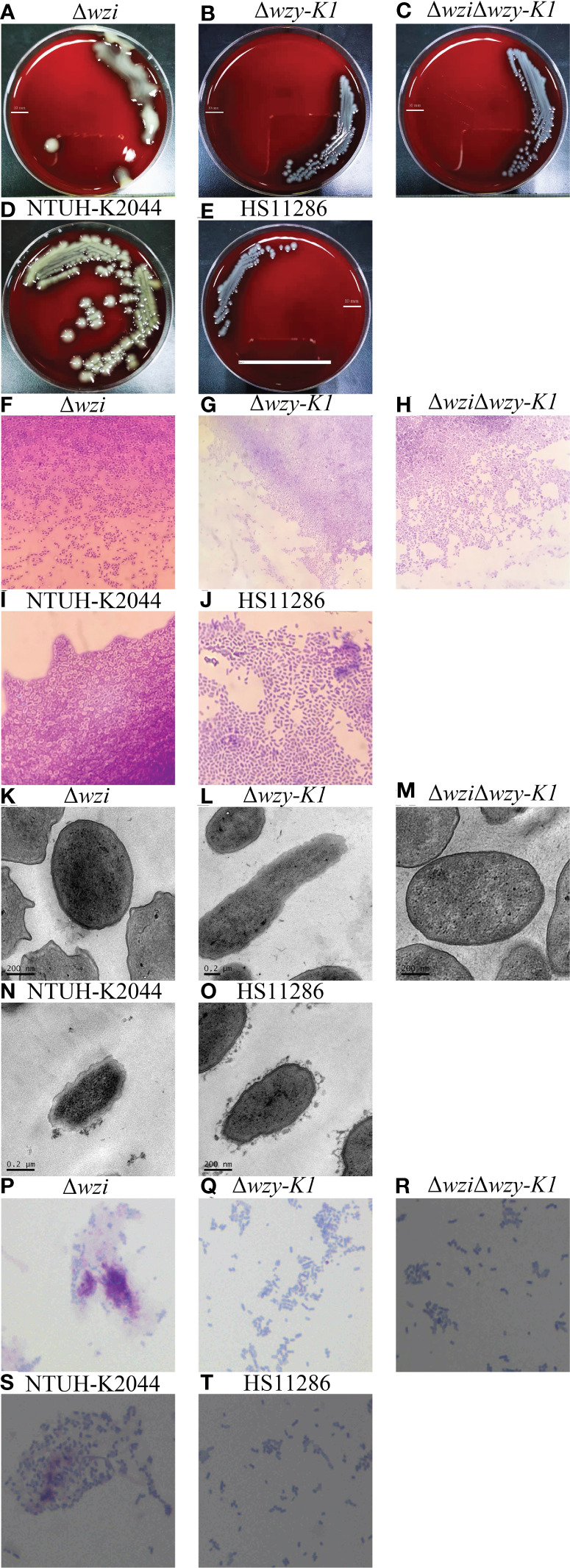
Morphological effects of deletions of wzi, wzy-K1, and wzi+wzy-K1 on Klebsiella pneumoniae. (A–E) Colonies of Δwzi, Δwzy-K1, ΔwziΔwzy-K1, NTUH-K2044, and HS11286; (F–J) Capsule staining of Δwzi, Δwzy-K1, ΔwziΔwzy-K1, NTUH-K2044, and HS11286; (K–O) Transmission electron microscopy of Δwzi, Δwzy-K1, ΔwziΔwzy-K1, NTUH-K2044, and HS11286; (P–T) Periodic acid-Schiff staining of Δwzi, Δwzy-K1, ΔwziΔwzy-K1, NTUH-K2044, and HS11286. The lines in (A–E) represent 10 mm; K. pneumoniae strains are purple and rod-shaped, and their transparent surroundings are hypercapsules (×1000) (F–J). The prominent “black edge” on the edge of the cell is the cell wall, and the loose material outside is the capsule (K–O). K. pneumoniae strains are blue and rod-shaped; the red fluffy masses are exopolysaccharides (P–T). The diameters of Δwzi, Δwzy-K1, ΔwziΔwzy-K1, NTUH-K2044, and HS11286 fell within such ranges: 8.0-10.0, 6.0-8.0, 2.0-4.0, 2.0-4.0, and 2.0-4.0 mm (A–E).
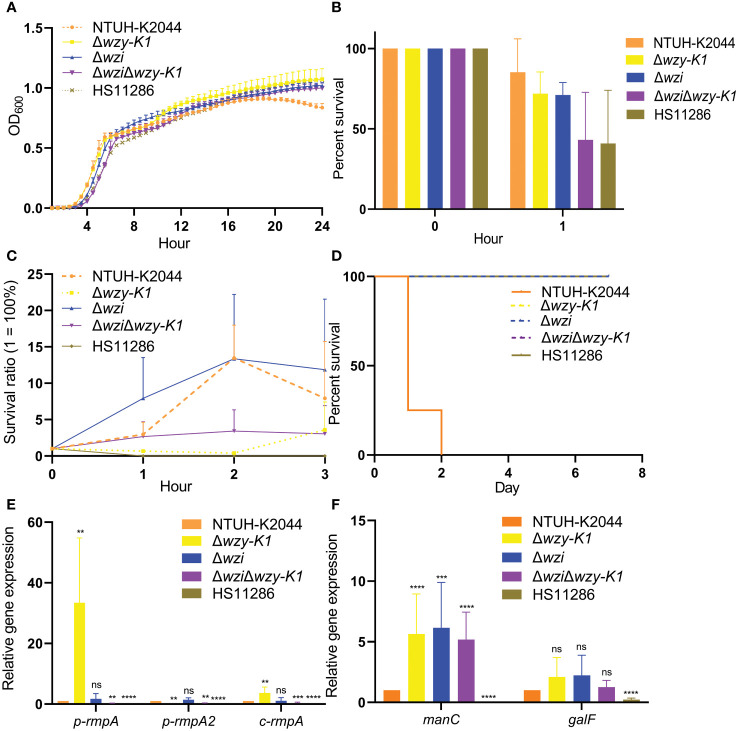
Growth curves showed no significant differences among NTUH-K2044, Δwzi, Δwzy-K1, ΔwziΔwzy-K1, and HS11286 ( Figure 3A ). The survival percentages were > 70.0% for Δwzi and Δwzy-K1 in the human neutrophil killing assay, while they decreased to ~40.0% for ΔwziΔwzy-K1 and HS11286 ( Figure 3B ). No significant differences were found among NTUH-K2044, Δwzi, Δwzy-K1, and ΔwziΔwzy-K1 for serum resistance ( Figure 3C ). Δwzi, Δwzy-K1, and ΔwziΔwzy-K1 all lost hypervirulence in the mouse lethality test ( Figure 3D ).
Figure 3.
Non-morphological effects of deletions of wzi, wzy-K1, and wzi+wzy-K1 on Klebsiella pneumoniae. (A) Growth curves; (B) Survival percentages in human neutrophil killing assay; (C) Survival ratios in serum killing assay; (D) Survival curves of mice infected with knockouts; (E) Relative expression of p-rmpA, p-rmpA2, and c-rmpA in knockouts; (F) Relative expression of manC and galF in knockouts. OD600: optical density at 600 nm; ns: not significant; **p < 0.01; ***p < 0.001; ****p < 0.0001. NTUH-K2044 and HS11286 were used as positive and negative controls, respectively. One-way ANOVA showed no significant differences among the 5 groups (A) (F = 0.0455, p = 0.9871). The values of means and standard deviations were 85.20/20.83, 71.90/13.53, 71.07/7.73, 43.17/29.60, and 40.83/33.18 respectively for NTUH-K2044, Δwzy-K1, Δwzi, ΔwziΔwzy-K1, and HS11286 after 1-h incubation (B). Homogeneity test of variance showed p = 0.0005 using Levene’s test. Kruskal-Wallis test presented a p value of 0.0065 among the 5 groups; while HS11286 was omitted, p = 0.1169 among the other 4 groups (C). The LD50 value of 106 CFU were for NTUH-K2044 in mouse lethality test, of which was >107 CFU for other 4 groups. The inoculation of 106 CFU was performed for the survival curve. Log-rank (Mantel-Cox) test showed χ2 = 23.0252 and p = 0.0001 among the 5 groups. Except NTUH-K2044, the other 4 groups showed the same survival curve; their comparison showed χ2 = 7.6037, p = 0.0058 (D). The relative expressions of p-rmpA, p-rmpA2, and c-rmpA were compared with those in NTUH-K2044. For the expression of p-rmpA, the means and standard deviations of △CT values were 19.52/0.87, 14.81/1.38, 19.13/1.07, 22.10/0.73, and 27.62/2.53 respectively in NTUH-K2044, Δwzy-K1, Δwzi, ΔwziΔwzy-K1, and HS11286; for the expression of p-rmpA2, such values were 16.03/0.82, 25.88/2.33, 15.72/1.49, 19.92/2.49, and 25.18/2.36; for the expression of c-rmpA, such values were 16.83/1.80, 15.19/2.59, 17.31/2.56, 19.29/1.38, and 26.51/1.76 (E). The relative expressions of manC and galF were compared with those in NTUH-K2044. For the expression of manC, the means and standard deviations of △CT values were 18.62/0.95, 30.47/1.86, 16.89/1.88, 16.58/0.60, and 27.76/1.26 respectively in NTUH-K2044, Δwzy-K1, Δwzi, ΔwziΔwzy-K1, and HS11286; for the expression of galF, such values were 13.06/1.05, 16.79/0.69, 12.51/0.78, 13.33/0.81, and 15.36/0.88 (F).
Figure 3E shows lower expression of p-rmpA, p-rmpA2, and c-rmpA in Δwzi, Δwzy-K1, and ΔwziΔwzy-K1 than in NTUH-K2044. Figure 3F shows equal expression of galF and higher expression of manC in Δwzi, Δwzy-K1, and ΔwziΔwzy-K1 than in NTUH-K2044.
Impact of CPS and EPS on cytokine secretion
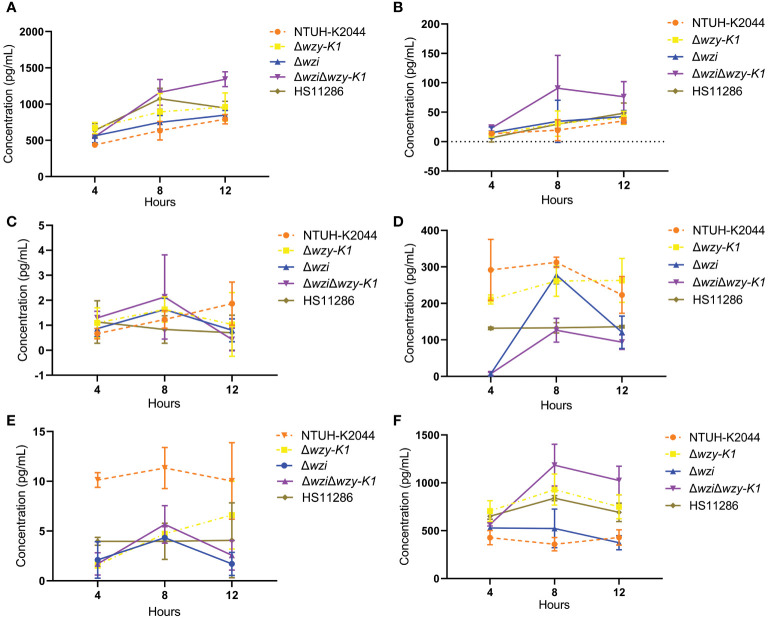
No significant differences were found among Δwzi, Δwzy-K1, ΔwziΔwzy-K1, and NTUH-K2044 for IL-6, IL-12, IL-10, and TGF-β secretions ( Figures 4A–D ) in the Kupffer cell stimulation assay. The secretion of IL-1β was lower for Δwzi, Δwzy-K1, and ΔwziΔwzy-K1 than for NTUH-K2044 ( Figure 4E ), which was the opposite of that observed with TNF-α ( Figure 4F ).
Figure 4.
Effects of deletions of wzi, wzy-K1, and wzi+wzy-K1 in Klebsiella pneumoniae on cytokine secretion by Kupffer cells. (A) IL-6 secretion; (B) IL-12 secretion; (C) IL-10 secretion; (D) TGF-β secretion; (E) IL-1β secretion; (F) TNF-α secretion. IL: interleukin; TGF-β: transforming growth factor-beta; TNF-α: tumor necrosis factor-alpha. NTUH-K2044 and HS11286 were used as positive and negative controls, respectively. Homogeneity test of variance showed p = 0.0106 using Levene’s test. Kruskal-Wallis test presented a p value of 0.4576 among the 5 groups (A). The factorial design analysis of variance showed significant differences among the 5 groups: F = 9.7138, p = 0.0018; further one-one comparisons confirmed all p values > 0.05; Comparison of each time point suggested F = 9.0737 and p = 0.0092. Analysis of interaction between groups and time points showed F = 0.8345, p = 0.5834 (B). ANOVA showed no significant differences among the 5 groups: F = 0.7462, p = 0.5823; Analysis of interaction between groups and time points showed F = 1.1687, p = 0.3644 (C). Normality test showed p = 0.0192 using Kolmogorov-Smirnov test. Kruskal-Wallis test presented a p value of 0.0264 among the 5 groups; further one-one comparisons confirmed all p values > 0.05 (D). ANOVA showed significant differences among the 5 groups: F = 38.9224, p < 0.0001 with NTUH-K2044 being higher than all the other 4 groups; the latter 4 groups all equalled: p > 0.05. Analysis of interaction between groups and time points showed F = 1.1674, p = 0.3651 (E). ANOVA showed significant differences among the 5 groups: F = 36.4485, p < 0.0001; further one-one comparisons confirmed such comparisons with p values > 0.05: NTUH-K2044 vs. Δwzi、Δwzy-K1 vs. ΔwziΔwzy-K1、Δwzy-K1 vs. HS11286、ΔwziΔwzy-K1 vs. HS11286, the others being with p < 0.05. Analysis of interaction between groups and time points showed F = 4.8494, p = 0.0020 (F).
Discussion
This investigation showed the differences between K. pneumoniae strains isolated from PLA and non-PLA samples; the roles of CPS and EPS in PLA; and the effects of CPS and EPS on cytokine secretion. Mutants of NTUH-K2044 were constructed through the deletions of wzy-K1, wzi, and the both. The gene wzy-K1 is a key virulence gene in K1 K. pneumoniae, encoding the Wzx/Wzy channel that is crucial for the synthesis of CPS and EPS (Schmid et al., 2015). The gene wzi encodes Wzi, which is the only anchor for CPS (Paczosa and Mecsas, 2016).
Figure 1 shows the differences between K. pneumoniae strains isolated from PLA and non-PLA samples. PLA is commonly community-acquired while a high proportion of non-PLA is hospital-acquired; Figures 1C , D confirmed such differences: ST23/ST65/ST700/ST660 and K1/K2 were widely found in K. pneumoniae strains from PLA while ST11 and K47 accounted for the majority in those from non-PLA. The proportion of serotype K1 in K. pneumoniae from PLA declined to 46.5% (20/43) compared with that in a previous report from five to nine years ago (68.9%, 31/45) (χ2 = 4.5186, p = 0.0335) (Qu et al., 2015), while the proportion of K2 was equal (9/45 vs. 10/43) (χ2 = 0.1377, p = 0.7106). The proportion of ST23 also declined from 57.8% (26/45) to 30.2% (13/43) (χ2 = 6.761, p = 0.0093). Nevertheless, K1 still dominates the serotypes of K. pneumoniae strains from PLA. Another study (Yu et al., 2006) showed that wzy-K1 could confer hypercapsule production in the absence of rmpA and rmpA2. Nearly half of the K. pneumoniae strains from PLA harboured wzy-K1, suggesting its important role in PLA. Based on positive wzy-K1, gene c-rmpA was the only difference between K. pneumoniae from PLA and non-PLA samples, which indicates a possible role of c-rmpA in PLA. The mere existence of c-rmpA in strains from PLA suggests its high specificity to PLA. Serotypes of K. pneumoniae strains could be determined by such methods: serological test and regular PCR to detect wzy-K1、PCR amplification and sequencing of wzi (Fang et al., 2007; Yeh et al., 2007; Turton et al., 2010). Usually, the results of such three methods show extremely high consistency. Gene wzy-K1 is thought to be in line with K1 K. pneumoniae (Struve et al., 2005; Yeh et al., 2006; Fang et al., 2010). Figure 1H presented some inconsistency. In this study, the last method was used to define serotypes. Presumably, mutations of wzi caused the inconsistency.
Figures 2F, K confirm the disappearance of the capsule in Δwzi. Compared to NTUH-K2044, the larger colonies of Δwzi ( Figure 2A ) resulted from the continuous synthesis of EPS and ropy CPS. Figures 2G , 2L , and 2Q verified the thin capsule and disappearance of EPS in Δwzy-K1, which resulted in smaller colonies ( Figure 2B ) compared to those of NTUH-K2044. The disappearance of both CPS and EPS was confirmed in ΔwziΔwzy-K1 ( Figures 2H, M, R ), which also resulted in smaller colonies ( Figure 2C ). NTUH-K2044 harbours 4 siderophores, i.e. aerobactin, salmochelin, yersiniabactin, and enterobactin. All three deletions showed no fitness cost ( Figure 3A ) and the four siderophores were retained. Virulence genes manC and galF are located at the downstream and upstream of cps cluster, which are often used to indicate the synthesis of CPS (Palacios et al., 2018; Peng et al., 2018). The higher expression of p-rmpA, c-rmpA, and manC in Δwzy-K1 resulted from the continuous synthesis of CPS precursors and blocking of the Wzx/Wzy channel ( Figures 3E, F ). With a functional Wzx/Wzy channel, normal expression of rmpAs was found in Δwzi; ΔwziΔwzy-K1 resulted in decreased expression of rmpAs. No significant impact of serum resistance was found in Δwzi, Δwzy-K1, and ΔwziΔwzy-K1 ( Figure 3C ). The survival rates were > 70.0% for Δwzi and Δwzy-K1 in the human neutrophil killing assay, while it was reduced to ~40.0% for ΔwziΔwzy-K1 ( Figure 3B ), indicating the role of EPS and thin capsules in protection against neutrophils. Figure 3D confirms the hypovirulence of Δwzi, Δwzy-K1, and ΔwziΔwzy-K1, although they all possessed four siderophores, suggesting that the hypercapsule is the core factor of hypervirulence rather than EPS.
Kupffer cells in the liver can secrete large amounts of proinflammatory cytokines, such as TNF-α, IL-1β, IL-6, and IL-12, and chemokines, such as C-X-C motif chemokine ligand 1 (CXCL1), CXCL2, CXCL3, CXCL8, and CXCL4 when exposed to bacteria (Liaskou et al., 2012). However, many anti-inflammatory cytokines are synthesised, such as IL-10 and TGF-β, under normal conditions. Figure 4 shows differences in IL-1β and TNF-α, but not IL-6, IL-12, IL-10, or TGF-β secretion in Δwzi, Δwzy-K1, and ΔwziΔwzy-K1 versus NTUH-K2044. IL-1β decreased with all three deletions ( Figure 4E ). The trend for TNF-α was not the same versus NTUH-K2044, increasing with Δwzy-K1 and ΔwziΔwzy-K1 but equal with Δwzi ( Figure 4F ). Figure 4 indicates that EPS in NTUH-K2044 contributes to immune escape from macrophages and attenuates the immune response of Kupffer cells, which is consistent with previous reports (Yoshida et al., 2000; Yoshida et al., 2001). The decreased IL-1β production of Δwzi, Δwzy-K1, and ΔwziΔwzy-K1, equal to that of HS11286 ( Figure 4E ), reflects the roles of CPS and EPS in HvKP, inducing the secretion of IL-1β from Kupffer cells and consequent hepatic lesions. The manifestations of TNF-α and IL-1β are likely to be contradictory. However, TNF-α plays a more important role in the process of PLA (Li et al., 2014). HS11286 harbours yersiniabactin and enterobactin according to Table 1 . Therefore, Δwzy-K1 shows traits similar to those of HS11286: thin capsule, no EPS, and types 1 and 3 fimbriae. The crucial difference between Δwzy-K1 and HS11286 is the numbers of siderophores: four vs. two. Figure 4 confirms equal secretion of the six cytokines between Δwzy-K1 and HS11286, which suggests the inability of excessive siderophores to induce inflammatory responses in Kupffer cells.
This study has some limitations. First, the sample size of strains from PLA is small, which may bring bias in Figure 1 . Second, the PLA mouse model is difficult to establish because of the hypovirulence of Δwzi, Δwzy-K1, and ΔwziΔwzy-K1. The majority of methods was then conducted in vitro in our study.
Conclusions
In summary, the presence of a hypercapsule is the cornerstone of hypervirulence regardless of the presence of EPS. The strategy of K1 K. pneumoniae in inducing PLA is to decrease the expression of core inflammatory cytokines rather than increase expression of anti-inflammatory cytokines. In contrast with CPS, EPS can effectively reduce the inflammatory response to aid in the immune escape of K. pneumoniae.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://pan.baidu.com/s/1iL6WDemsLyPJepNNrYbTiQ?pwd=1234; Key: 1234.
Ethics statement
The studies involving human participants were reviewed and approved by the Ethics Committee of Huashan Hospital (Shanghai, China) (ethical approval No. 2021-484). Written informed consent for participation was not required for this study in accordance with the national legislation and the institutional requirements. The animal study was reviewed and approved by the Institutional Animal Care and Use Committee of the School of Pharmacy, Fudan University (Ethical approval No. 201603-TY-MQ-01).
Author contributions
DH, WC, and WW conceived the study. GL, and XJ collected and identified the strains. DH, WC, WW, PF, and DT performed PCR and MLST analyses, string tests, capsular staining, periodic acid–Schiff staining, ELISA, gene deletion, Galleria mellonella lethality and fitness tests. PR and QM performed mouse lethality tests. DH, WC, and WW wrote the manuscript which was revised by XJ and GL. All authors contributed to the article and approved the submitted version.
Acknowledgments
We thank Professor Jin-Town Wang from the Department of Internal Medicine, National Taiwan University Hospital for authorizing the strain NTUH-K2044. We also thank such researchers for collecting Klebsiella pneumoniae strains: Meng Li (Department of Clinical Laboratory, the First Affiliated Hospital of Guangxi Medical University, Nanning 530021, Guangxi, China), Zehua Yang (Department of Laboratory Medicine, Sixth Hospital of Shanxi Medical University, Taiyuan 030008, Shanxi, China), Yong Wang (Department of Clinical Laboratory, Shandong Provincial Hospital Affiliated to Shandong University, Jinan 250021, Shandong, China), Yunkun Huang (Department of Laboratory Medicine, Kunming Yan’an Hospital, Kunming 650051, Yunnan, China) and Lianhua Yu (Department of Laboratory Medicine, Taizhou Municipal Hospital, Taizhou 318000, Zhejiang, China).
Funding Statement
This study was funded by research grants from the National Natural Science Foundation of China (grants 81871692 and 81572031), Shanghai Municipal Key Clinical Specialty (Laboratory Medicine, grant No. shslczdzk03303), and the Shanghai Municipal Science and Technology Commission (grant No. 19JC1413002). The fundings have no role in study design, data collection, the writing of the manuscript and decision to submit it for publication.
Abbreviations
PLA, pyogenic liver abscess; K. pneumoniae, Klebsiella pneumoniae; HvKP, hypervirulent K. pneumoniae; cKP, classical K. pneumoniae; EPS, Exopolysaccharide; CPS, capsular polysaccharide; rmpA, regulator of mucoid phenotype A gene; MLST, multilocus sequence typing; ST, sequence type; CFU, colony forming unit; TEM, transmission electron microscopy; PBS, phosphate buffered saline; LB, Luria Bertani; OD, optical density; LD50, lethality dose 50; G. mellonella, Galleria mellonella; TNF-α, tumor-necrosis factor-α; TGF-β, transforming growth factor-β; ST, sequence type; CXCL1, C-X-C motif chemokine ligand 1; ns, not significant; ND, not defined.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2023.1147855/full#supplementary-material
References
- Compain F., Babosan A., Brisse S., Genel N., Audo J., Ailloud F., et al. (2014). Multiplex PCR for detection of seven virulence factors and K1/K2 capsular serotypes of klebsiella pneumoniae. J. Clin. Microbiol. 52, 4377–4380. doi: 10.1128/JCM.02316-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko K. A., Wanner B. L. (2000). One-step inactivation of chromosomal genes in escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 97, 6640–6645. doi: 10.1073/pnas.120163297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deleo F. R., Chen L., Porcella S. F., Martens C. A., Kobayashi S. D., Porter A. R., et al. (2014). Molecular dissection of the evolution of carbapenem-resistant multilocus sequence type 258 klebsiella pneumoniae. Proc. Natl. Acad. Sci. U.S.A. 111, 4988–4993. doi: 10.1073/pnas.1321364111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst C. M., Braxton J. R., Rodriguez-Osorio C. A., Zagieboylo A. P., Li L., Pironti A., et al. (2020). Adaptive evolution of virulence and persistence in carbapenem-resistant klebsiella pneumoniae. Nat. Med. 26, 705–711. doi: 10.1038/s41591-020-0825-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang C. T., Lai S. Y., Yi W. C., Hsueh P. R., Liu K. L. (2010). The function of wzy_K1 (magA), the serotype K1 polymerase gene in klebsiella pneumoniae cps gene cluster. J. Infect. Dis. 201, 1268–1269. doi: 10.1086/652183 [DOI] [PubMed] [Google Scholar]
- Fang C. T., Lai S. Y., Yi W. C., Hsueh P. R., Liu K. L., Chang S. C. (2007). Klebsiella pneumoniae genotype K1: An emerging pathogen that causes septic ocular or central nervous system complications from pyogenic liver abscess. Clin. Infect. Dis. 45, 284–293. doi: 10.1086/519262 [DOI] [PubMed] [Google Scholar]
- Gu D., Dong N., Zheng Z., Lin D., Huang M., Wang L., et al. (2018). A fatal outbreak of ST11 carbapenem-resistant hypervirulent klebsiella pneumoniae in a Chinese hospital: a molecular epidemiological study. Lancet Infect. Dis. 18, 37–46. doi: 10.1016/S1473-3099(17)30489-9 [DOI] [PubMed] [Google Scholar]
- Hoh C. H., Tan Y. H., Gan Y. H. (2019). Protective role of kupffer cells and macrophages in klebsiella pneumoniae-induced liver abscess disease. Infect. Immun. 87, e00369-19. doi: 10.1128/IAI.00369-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalitha C., Raman T., Rathore S. S., Ramar M., Munusamy A., Ramakrishnan J. (2017). ASK2 bioactive compound inhibits MDR klebsiella pneumoniae by antibiofilm activity, modulating macrophage cytokines and opsonophagocytosis. Front. Cell Infect. Microbiol. 7, 346. doi: 10.3389/fcimb.2017.00346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Zhao Y., Liu C., Chen Z., Zhou D. (2014). Molecular pathogenesis of klebsiella pneumoniae. Future Microbiol. 9, 1071–1081. doi: 10.2217/fmb.14.48 [DOI] [PubMed] [Google Scholar]
- Liaskou E., Wilson D. V., Oo Y. H. (2012). Innate immune cells in liver inflammation. Mediators Inflammation 2012, 949157. doi: 10.1155/2012/949157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J. C., Koh T. H., Lee N., Fung C. P., Chang F. Y., Tsai Y. K., et al. (2014). Genotypes and virulence in serotype K2 klebsiella pneumoniae from liver abscess and non-infectious carriers in Hong Kong, Singapore and Taiwan. Gut Pathog. 6, 21. doi: 10.1186/1757-4749-6-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y. T., Siu L. K., Lin J. C., Chen T. L., Tseng C. P., Yeh K. M., et al. (2012). Seroepidemiology of klebsiella pneumoniae colonizing the intestinal tract of healthy Chinese and overseas Chinese adults in Asian countries. BMC Microbiol. 12, 13. doi: 10.1186/1471-2180-12-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D., Liu Z. S., Hu P., Cai L., Fu B. Q., Li Y. S., et al. (2016). Characterization of surface antigen protein 1 (SurA1) from acinetobacter baumannii and its role in virulence and fitness. Vet. Microbiol. 186, 126–138. doi: 10.1016/j.vetmic.2016.02.018 [DOI] [PubMed] [Google Scholar]
- Lo J. Z., Leow J. J., Ng P. L., Lee H. Q., Mohd Noor N. A., Low J. K., et al. (2015). Predictors of therapy failure in a series of 741 adult pyogenic liver abscesses. J. Hepatobiliary Pancreat Sci. 22, 156–165. doi: 10.1002/jhbp.174 [DOI] [PubMed] [Google Scholar]
- Meddings L., Myers R. P., Hubbard J., Shaheen A. A., Laupland K. B., Dixon E., et al. (2010). A population-based study of pyogenic liver abscesses in the united states: incidence, mortality, and temporal trends. Am. J. Gastroenterol. 105, 117–124. doi: 10.1038/ajg.2009.614 [DOI] [PubMed] [Google Scholar]
- Mizuta K., Ohta M., Mori M., Hasegawa T., Nakashima I., Kato N. (1983). Virulence for mice of klebsiella strains belonging to the O1 group: relationship to their capsular (K) types. Infect. Immun. 40, 56–61. doi: 10.1128/iai.40.1.56-61.1983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paczosa M. K., Mecsas J. (2016). Klebsiella pneumoniae: Going on the offense with a strong defense. Microbiol. Mol. Biol. Rev. 80, 629–661. doi: 10.1128/MMBR.00078-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacios M., Miner T. A., Frederick D. R., Sepulveda V. E., Quinn J. D., Walker K. A., et al. (2018). Identification of two regulators of virulence that are conserved in klebsiella pneumoniae classical and hypervirulent strains. mBio 9, e01443-18. doi: 10.1128/mBio.01443-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng D., Li X., Liu P., Zhou X., Luo M., Su K., et al. (2018). Transcriptional regulation of galF by RcsAB affects capsular polysaccharide formation in klebsiella pneumoniae NTUH-K2044. Microbiol. Res. 216, 70–78. doi: 10.1016/j.micres.2018.08.010 [DOI] [PubMed] [Google Scholar]
- Podschun R., Pietsch S., Holler C., Ullmann U. (2001). Incidence of klebsiella species in surface waters and their expression of virulence factors. Appl. Environ. Microbiol. 67, 3325–3327. doi: 10.1128/AEM.67.7.3325-3327.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu T. T., Zhou J. C., Jiang Y., Shi K. R., Li B., Shen P., et al. (2015). Clinical and microbiological characteristics of klebsiella pneumoniae liver abscess in East China. BMC Infect. Dis. 15, 161. doi: 10.1186/s12879-015-0899-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed L. J., Muench H. (1938). A simple method of estimating fifty percent endpoints. Am. J. Hyg. 27, 493–497. [Google Scholar]
- Roediger R., Lisker-Melman M. (2020). Pyogenic and amebic infections of the liver. Gastroenterol. Clin. North Am. 49, 361–377. doi: 10.1016/j.gtc.2020.01.013 [DOI] [PubMed] [Google Scholar]
- Russo T. A., Marr C. M. (2019). Hypervirulent klebsiella pneumoniae. Clin. Microbiol. Rev. 32, e00001-19. doi: 10.1128/CMR.00001-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo T. A., Olson R., Fang C. T., Stoesser N., Miller M., MacDonald U., et al. (2018). Identification of biomarkers for differentiation of hypervirulent klebsiella pneumoniae from classical k. pneumoniae. J. Clin. Microbiol. 56, e00776-18. doi: 10.1128/JCM.00776-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid J., Sieber V., Rehm B. (2015). Bacterial exopolysaccharides: biosynthesis pathways and engineering strategies. Front. Microbiol. 6, 496. doi: 10.3389/fmicb.2015.00496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi S. H., Feng X. N., Lai M. C., Kong H. S., Zheng S. S. (2017). Biliary diseases as main causes of pyogenic liver abscess caused by extended-spectrum beta-lactamase-producing enterobacteriaceae. Liver Int. 37, 727–734. doi: 10.1111/liv.13267 [DOI] [PubMed] [Google Scholar]
- Shon A. S., Bajwa R. P., Russo T. A. (2013). Hypervirulent (hypermucoviscous) klebsiella pneumoniae: a new and dangerous breed. Virulence 4, 107–118. doi: 10.4161/viru.22718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struve C., Bojer M., Nielsen E. M., Hansen D. S., Krogfelt K. A. (2005). Investigation of the putative virulence gene magA in a worldwide collection of 495 klebsiella isolates: magA is restricted to the gene cluster of klebsiella pneumoniae capsule serotype K1. J. Med. Microbiol. 54, 1111–1113. doi: 10.1099/jmm.0.46165-0 [DOI] [PubMed] [Google Scholar]
- Tian L. T., Yao K., Zhang X. Y., Zhang Z. D., Liang Y. J., Yin D. L., et al. (2012). Liver abscesses in adult patients with and without diabetes mellitus: an analysis of the clinical characteristics, features of the causative pathogens, outcomes and predictors of fatality: a report based on a large population, retrospective study in China. Clin. Microbiol. Infect. 18, E314–E330. doi: 10.1111/j.1469-0691.2012.03912.x [DOI] [PubMed] [Google Scholar]
- Tsai F. C., Huang Y. T., Chang L. Y., Wang J. T. (2008). Pyogenic liver abscess as endemic disease, Taiwan. Emerg. Infect. Dis. 14, 1592–1600. doi: 10.3201/eid1410.071254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turton J. F., Perry C., Elgohari S., Hampton C. V. (2010). PCR characterization and typing of klebsiella pneumoniae using capsular type-specific, variable number tandem repeat and virulence gene targets. J. Med. Microbiol. 59, 541–547. doi: 10.1099/jmm.0.015198-0 [DOI] [PubMed] [Google Scholar]
- Yeh K. M., Chang F. Y., Fung C. P., Lin J. C., Siu L. K. (2006). magA is not a specific virulence gene for klebsiella pneumoniae strains causing liver abscess but is part of the capsular polysaccharide gene cluster of k. pneumoniae serotype K1. J. Med. Microbiol. 55, 803–804. doi: 10.1099/jmm.0.46368-0 [DOI] [PubMed] [Google Scholar]
- Yeh K. M., Kurup A., Siu L. K., Koh Y. L., Fung C. P., Lin J. C., et al. (2007). Capsular serotype K1 or K2, rather than magA and rmpA, is a major virulence determinant for klebsiella pneumoniae liver abscess in Singapore and Taiwan. J. Clin. Microbiol. 45, 466–471. doi: 10.1128/JCM.01150-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida K., Matsumoto T., Tateda K., Uchida K., Tsujimoto S., Yamaguchi K. (2000). Role of bacterial capsule in local and systemic inflammatory responses of mice during pulmonary infection with klebsiella pneumoniae. J. Med. Microbiol. 49, 1003–1010. doi: 10.1099/0022-1317-49-11-1003 [DOI] [PubMed] [Google Scholar]
- Yoshida K., Matsumoto T., Tateda K., Uchida K., Tsujimoto S., Yamaguchi K. (2001). Induction of interleukin-10 and down-regulation of cytokine production by klebsiella pneumoniae capsule in mice with pulmonary infection. J. Med. Microbiol. 50, 456–461. doi: 10.1099/0022-1317-50-5-456 [DOI] [PubMed] [Google Scholar]
- Yu W. L., Ko W. C., Cheng K. C., Lee H. C., Ke D. S., Lee C. C., et al. (2006). Association between rmpA and magA genes and clinical syndromes caused by klebsiella pneumoniae in Taiwan. Clin. Infect. Dis. 42, 1351–1358. doi: 10.1086/503420 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://pan.baidu.com/s/1iL6WDemsLyPJepNNrYbTiQ?pwd=1234; Key: 1234.