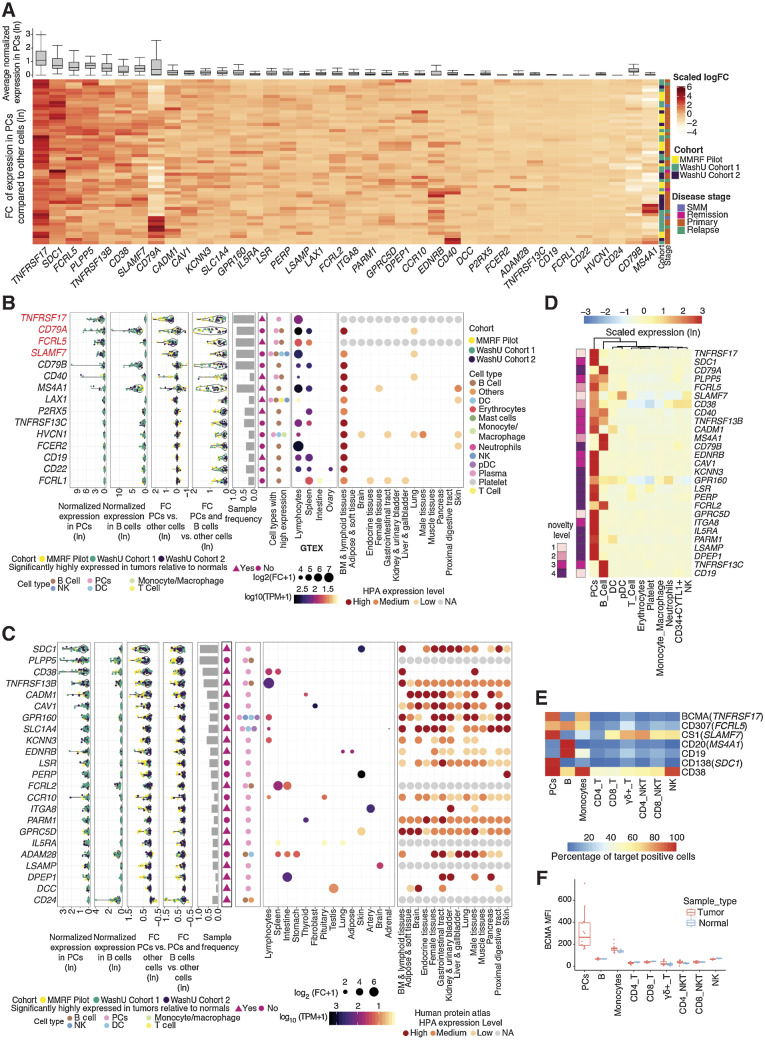
Figure 3.
Characterization of myeloma cell–associated surface proteins. A, Average gene expression in plasma cells (top, box and whisker plot) and hierarchical clustering of z-score scaled log-fold change between plasma and nonplasma cells across samples (bottom, heat map). B, Summarized characterization of primary candidate targets expressed on the cell surface with specific expression in lymphoid tissues. Genes are ordered by average expression in PCs. Top four genes are top-tier primary targets, highlighted in red. Average normalized expression in plasma and B cells are shown in the first two sections, respectively. Average expression log-fold change between PCs and other cells is shown in the third section, followed by expression log-fold change of combined plasma and B cells compared with other cells shown in the fourth section (FC, fold change). Each dot represents a sample, colored by its corresponding cohort. Sample frequency is the proportion of samples having genes differentially expressed in PCs or in combined plasma and B-cell population if the gene is only expressed in B cells. In the sixth section, triangle and circle denote whether genes are significantly highly expressed in tumors relative to normal BMs. Cell types with target expression are annotated in the seventh section. In the GTEx tissue expression specificity analysis section, only gene-tissue type relationships with a significant FDR are plotted. Size corresponds to fold difference and is colored by expression. HPA protein expression was shown in the last section, with color indicating expression level. NA, not available. C, Summarized characterization of secondary candidate targets expressed on the cell surface without specific expression in lymphoid tissues. Figure layout is the same as B. D, Heat map showing the z-score scaled average expression of known and novel targets across cell populations in scRNA-seq. Columns are ordered based on the hierarchical clustering of target expressions; rows are ordered by average normalized expression in plasma cells. Left annotation indicates novelty level: 1, currently under clinical study as CAR-based therapy; 2, currently under clinical study as antibody-based therapy; 3, potential therapeutic utility is supported by existing literature; 4, previously uncharacterized in its capacity as a myeloma marker. Further details for novelty ranking are included in Supplementary Table S2. E, Heat map showing the average percentage of cells with positive expression of surface antigens across cell populations from 12 patient samples (11 MM and 1 SMM patients) in flow cytometry. F, Box plot showing mean fluorescence intensity (MFI) of BCMA in patient BMMC (n = 12, including 11 MM and 1 SMM) or PBMC from healthy donors (n = 3) across cell populations in flow cytometry.