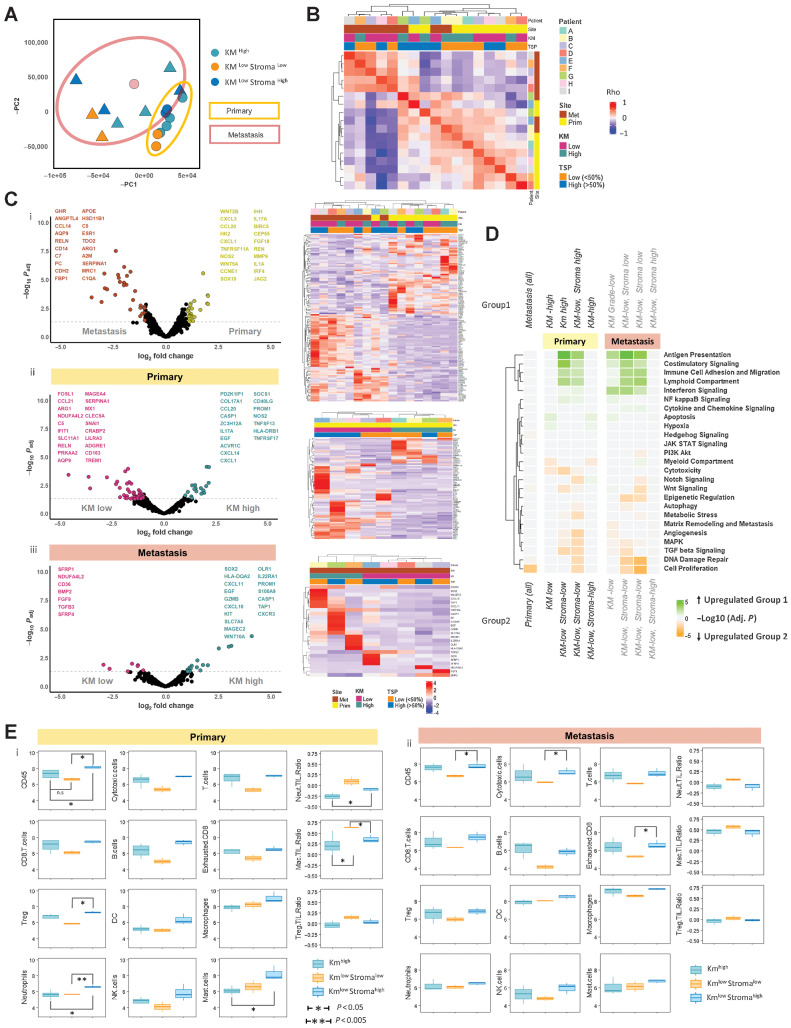
Figure 2.
BULK IO360 transcriptomic characterization of matched primary colorectal cancer and CRLM. A, PCA plot demonstrating two principal components of minimal variance for all samples. Primary colorectal cancer samples are demonstrated by circles and yellow outline. CRLM are demonstrated by triangular points and brown outline. KMhigh, KMlow Stromalow and KMlow Stromahigh samples are depicted by color. B, Unsupervised analysis using gene expression correlation matrix for all samples. Patient, site, KM grade, and TSP are depicted by key. Spearman correlation of all expressed genes performed between each sample sequenced and plotted on the heatmap. k-means clustering of heatmap to demonstrate correlated samples. Red, strong correlation. Blue, negative correlation. C, Volcano plots demonstrating differential gene expression results and clustered heatmap of significant genes for (i) all primary colorectal cancer versus CRLM; (ii) KM grade: KM high versus KM low primary colorectal cancer; (iii) KM grade: KM high versus KM low CRLM. The x-axis of volcano plot demonstrates log2-fold change; y-axis demonstrates –log10P. Colored points demonstrate significant changes in gene expression between groups (P < 0.05 and logFC > 1.5). Volcano plots demonstrate top 20 differentially expressed genes for each group. D, Heatmap demonstrating GSEA results comparing the different tumors grouped according with KM grade and TSP using io360-curated gene sets annotated on the right of the diagram. Heatmap squares represent log10-adjusted P value. Green, upregulation in group 1; orange, upregulation in group 2. The heatmap is clustered by y-axis only to demonstrate frequently upregulated gene sets. E, Box plot comparisons of immune cell populations and selected cell:cell ratios between KM grade and TSP segregated groups using deconvolution software included in the nCounter package. Annotated subgroups are: Kmhigh, Kmlow Stromalow, Kmlow Stromahigh. The y-axis represents log10 of estimated cell count. Primary colorectal cancer is represented in i and CRLM in ii.