Figure 5. Off-targets with single-nucleotide deletions are accommodated by base skipping or multiple consecutive mismatches.
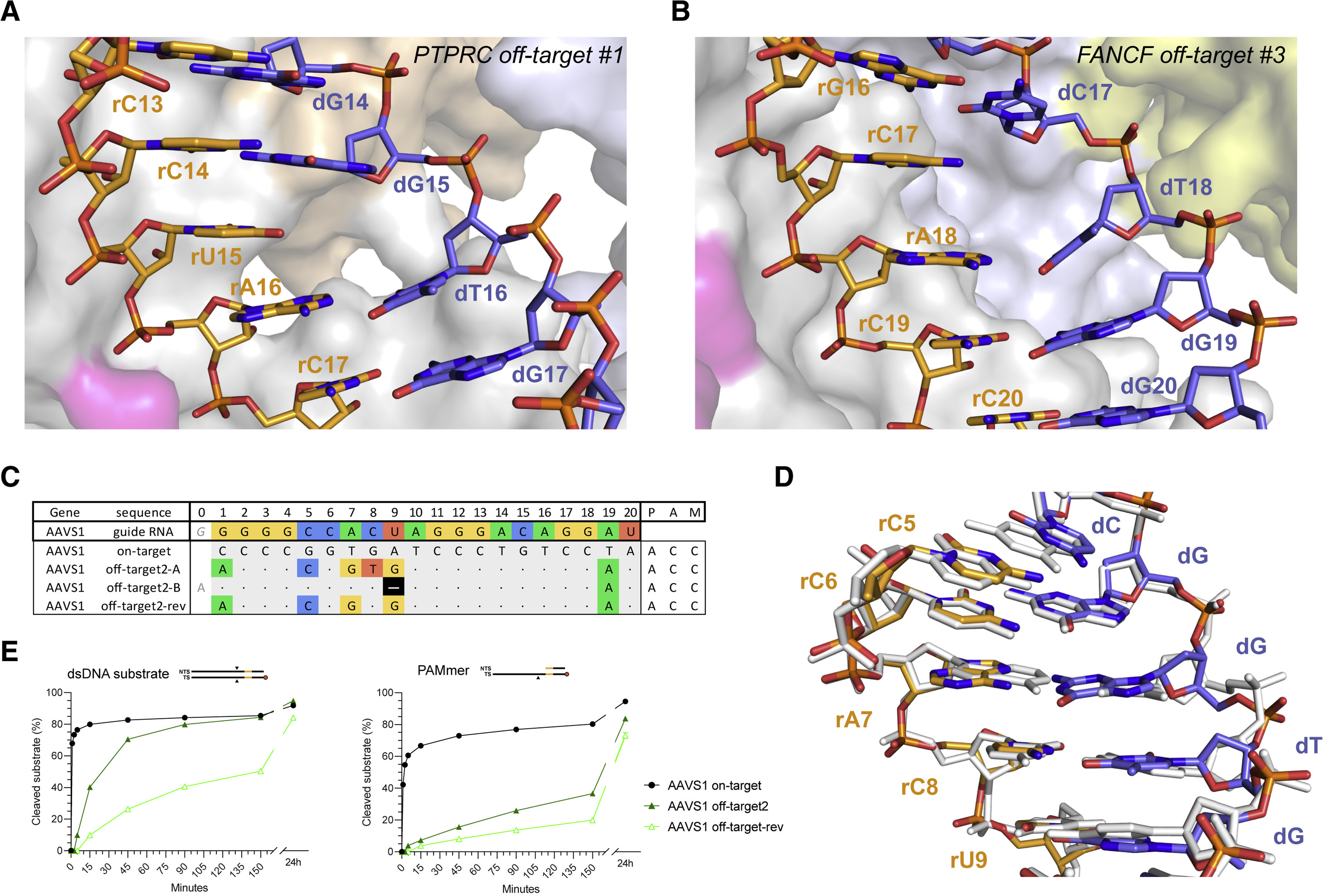
(A) Zoomed-in view of the base skip at duplex position 15 in the PTPRC-tgt2 off-target #1 complex. (B) Zoomed-in view of the base skip at duplex position 17 in the FANCF off-target #3 complex. (C) Schematic depiction of alternative base pairing interactions in the AAVS1 off-target #2 complex. AAVS1 off-target #2-rev substrate was designed based on the AAVS1 off-target #2, with the reversal of a single mismatch in the consecutive region back to the corresponding canonical base pair. (D) Structural overlay of the AAVS1 off-target #2 (coloured) and AAVS1 on-target (white) heteroduplexes. (E) Cleavage DNA kinetics of AAVS1 on-target, off-target #2 and off-target #2-rev substrates. See also Figures S15, S16, S17, S18, S19.
