Abstract
CD8 cytotoxic T cells are a potent line of defense against invading pathogens. To aid in curtailing aberrant immune responses, the activation status of CD8 T cells is highly regulated. One mechanism in which CD8 T cell responses are dampened is via signaling through the immune-inhibitory receptor PD-1, encoded by Pdcd1. Pdcd1 expression is regulated through engagement of the T cell receptor as well as by signaling from extracellular cytokines. Understanding such pathways has influenced the development of numerous clinical treatments. Here, we showed that signals from the cytokine IL-6 enhanced Pdcd1 expression when paired with TCR stimulation in murine CD8 T cells. Mechanistically, signals from IL-6 were propagated through activation of the transcription factor STAT3, resulting in IL-6-dependent binding of STAT3 to Pdcd1 cis-regulatory elements. Intriguingly, IL-6 stimulation overcame BLIMP-1-mediated epigenetic repression of Pdcd1, which resulted in a transcriptionally permissive landscape marked by heightened histone acetylation. Furthermore, in vivo activated CD8 T cells derived from LCMV infection required STAT3 for optimal PD-1 surface expression. Importantly, STAT3 was the only member of the STAT family present at Pdcd1 regulatory elements in LCMV-antigen specific CD8 T cells. Collectively, these data define mechanisms by which the IL-6/STAT3 signaling axis can enhance and prolong Pdcd1 expression in murine CD8 T cells.
INTRODUCTION
A robust adaptive immune response requires contributions from CD8 cytotoxic T cells, which assist in viral pathogen and cancer cell clearance through direct killing of infected or cancerous cells (1–3). While the cytotoxic functions of CD8 T cells are pivotal to pathogen clearance, curtailing their function is crucial to preventing overactivity or autoimmune responses (4, 5). One such brake on aberrant CD8 T cell functions is the well documented upregulation of the surface protein Programmed Cell Death Protein-1 (PD-1), encoded by Pdcd1, which occurs following TCR stimulation (6, 7). Engagement of PD-1 by its ligand (PD-L1), results in a multitude of outcomes antagonistic to CD8 T cell activation, including blocking of co-stimulatory signals through CD28, decreased cytokine production, and initiating cell cycle arrest (8–13). The net outcome of CD8 T cells experiencing continued PD-1 signal transduction is a T cell state often referred to as exhaustion (14). As such, the PD-1 pathway has proven to be an effective target for cancer immunotherapies where blocking PD-1/PD-L1 interactions reinvigorates CD8 T cells to combat tumors (15–18).
Several studies have identified numerous cis- and trans-regulatory elements that control Pdcd1 expression (19–21). In response to TCR stimulation, NFATC1 binds to an element upstream of the transcription start site referred to as Conserved Region-C (CR-C) (19–22). Induction of the Pdcd1 locus is marked by changes in histone posttranslational modifications at key cis-regulatory elements, as well as changes in DNA methylation, with the known activities of these modifications each contributing to the regulation of the locus (20, 23, 24). Indeed, TCR stimulation of murine CD8 T cells is marked by loss of DNA methylation at CpGs, increased H3K27 acetylation (ac) and H3K9ac at CR-B (another cis-regulatory element close to the promoter) and CR-C, as well as enhanced formation of long-range interactions between distal regulatory regions and the Pdcd1 promoter (19, 23, 25, 26). In contrast, the transcription factor B-Lymphocyte Maturation Protein 1 (BLIMP-1), encoded by Prdm1, has been shown to repress Pdcd1 expression through the recruitment of the chromatin remodeling enzyme Lysine-Specific Demethylase 1a (LSD1). LSD1 reduces Pdcd1 expression through the decommissioning of enhancer and promoter elements in the Pdcd1 gene (26, 27).
Cytokines also influence the expression of Pdcd1 in CD8 T cells. IL-6 and IL-12 signaling through STAT3 and STAT4, respectively, have been shown to enhance TCR mediated induction of murine Pdcd1 (19, 28, 29). Signaling through these cytokines leads to JAK-dependent phosphorylation of their respective STAT factors, initiating STAT dimerization and translocation into the nucleus where they can augment gene expression (30–32). At the Pdcd1 locus, STAT3 has been shown to associate directly with key cis-regulatory elements, including the two enhancer elements located at −3.7kb and +17.1kb from the transcription start site (19). While such data begins to describe an IL-6-specific means of Pdcd1 regulation, questions remain regarding the exact, CD8 T cell-specific, mechanisms through which IL-6 promotes Pdcd1 expression.
This study focused on expanding the understanding of IL-6/STAT3-dependent regulation of Pdcd1 in murine CD8 T cells. Treatment of primary CD8 T cells with IL-6 led to enhanced and prolonged expression of Pdcd1, Stat3, and Prdm1. Furthermore, IL-6 signaling drove STAT3 phosphorylation/activation and subsequent binding to key Pdcd1 regulatory elements. STAT3 binding prevented BLIMP-1 from inducing a repressive epigenetic state while simultaneously promoting increased histone acetylation. Analysis of mice lacking STAT3 in activated CD8 T cells, revealed that IL-6 enhancement of Pdcd1 expression was dependent on STAT3 activities. Infection of STAT3-deficient mice with lymphocytic choriomeningitis virus (LCMV) clone 13, revealed a role of STAT3 in the induction of PD-1 expression in vivo. LCMV-derived CD8 T cells were enriched for STAT3 at several elements within the Pdcd1 locus. Collectively, these findings expand on IL-6-depenent regulation of Pdcd1 expression, detailing a requirement for STAT3 in optimal PD-1 expression in vivo.
MATERIALS AND METHODS
Cell culture
Murine EL4 cells were cultured in RPMI 1640 containing 5% FBS (Sigma-Aldrich), 5% bovine calf serum (HyClone, Inc.), 100 U/ml penicillin/streptomycin, 1.0 mM sodium pyruvate, 10 mM Hepes, and 4.5 mg/ml glucose. Primary murine CD8 T cells were isolated from spleens of C57BL/6 mice using a negative selection method with the CD8a+ T Cell Isolation kit II (Miltenyi Biotec) according to the manufacturer’s protocol. Newly isolated primary CD8 T cells were cultured in the same media as EL4 cells. Anti-CD3/CD28 beads (Invitrogen) were directly added to the media at a bead-to-cell ratio of 1:1 to activate the CD8 T cells. Cells were treated with 20 ng/ml IL-6 (Miltenyi Biotech) or 20 ng/ml IL-10 (Miltenyi Biotech).
Mice and P14 adoptive transfers
C57BL/6 WT, B6.129S1 (Stat3fl/fl), and B6;FVB-Tg1Jcb/J (GzmbCre) mice were purchased from Jackson Laboratories. STAT3 cKO mice were generated by breeding Stat3fl/fl mice with GzmbCre mice to delete STAT3 in activated T cells as previously described (26, 33). These are referred to as KO mice in the text. For some experiments, P14 mice were used (generously provided by Dr. Rafi Ahmed’s lab (Emory University)). P14 mice contain a TCR transgene for LCMV antigenic peptide GP33. For these experiments 10,000 P14 splenocytes were adoptively transferred into C57Bl/6 Thy1.2 mice via tail vein injection. Transferred cells were isolated from spleens of Thy1.2 hosts by positive magnetic selection kit on Thy1.1(Miltenyi Biotec). Purity of isolated cells was confirmed by flow cytometry for Thy1.1 expression. In other experiments, wild-type and STAT3 KO mice were used as indicated. For all experiments genotypes were confirmed by PCR. Mice used in this study were maintained and manipulated in compliance with the protocols approved by Emory University Institutional Animal Care and Use Committee.
ChIP Assays
Chromatin immunoprecipitation (ChIP) assays were performed as previously described (26). Briefly, except where noted, 1–4 × 107 cells were cross-linked in 1% formaldehyde for 15 min and then subjected to sonication to shear chromatin. 10 μg of chromatin was used for each immunoprecipitation. Chromatin was incubated with the indicated antibodies at 4º C overnight, then magnetic Protein A or Protein G beads (Invitrogen) were added to the sample and incubated at 4° C for 2 hours to isolate the chromatin-antibody complexes. Antibodies used for immunoprecipitation are listed in Supplemental Table 1. Non-immune rabbit IgG was used as a nonspecific control for all the rabbit antibodies. The mouse monoclonal antibody anti-HA (12CA5), which recognizes a short peptide from the influenza hemagglutinin protein, was used as a nonspecific control for the mouse monoclonal anti-NFATC1 antibody. The immunoprecipitated chromatin was eluted and incubated at 65° C overnight to reverse the protein-DNA cross-link, then chromatin DNA was purified and quantified by real-time PCR using a standard curve from sonicated murine genomic DNA. For H3K27ac ChIP on non-P14, LCMV-sorted CD8 T cells, 2–5 × 105 cells were used per immunoprecipitation. Because of the low cell input, Protein G beads were prebound with H3K27ac ab overnight before introducing sheared chromatin for an additional 24-hour incubation. Each ChIP assay was performed with chromatin purified from at least three independent experiments and represented relative to total input.
RNA extraction and qRT-PCR
RNA was isolated from primary murine CD8 T cells using the RNeasy Mini Kit (Qiagen, 74106) according to manufacturer’s instructions. Template DNA was digested with DNase for 30 min at 37° C. cDNA was generated using the Superscript II reverse transcriptase (Life Technologies, 18064–14). At least three-independent RNA preparations were used for real-time PCR analysis using site-specific primers for: Pdcd1, Prdm1, Stat3, and Stat4. All values were normalized to 18s rRNA. Primer sequences are found in Supplemental Table 1.
Immunoblot analysis
Cells were lysed on ice for 30 min in RIPA buffer (150 mmol/L NaCl, 50 mmol/L Tris (pH 7.4), 1% NP-40, and 0.5% Na-deoxycholate) with freshly added protease and phosphatase inhibitors and spun down at 4° C to remove debris. Protein concentrations were determined using Bradford protein assay (Bio-Rad). Cell lysates were resolved on 10% SDS-PAGE and transferred to polyvinylidene difluoride membranes. Antibodies used for blotting were listed in Supplemental Table 1. Protein band signals were detected with the ECL detection kit (Thermo-Fisher).
LCMV infection and titer measurement
Mice were infected with 2 × 106 plague forming units (PFU) of Lymphocytic choriomeningitis virus (LCMV) clone 13 i.v. as previously described to induce high PD-1 expression by CD8 T cells (27). All viral stocks were graciously provided by Dr. Rafi Ahmed (Emory University). WT and STAT3 cKO mice were infected in a blind fashion as to ensure equal virus administration across genotype. LCMV clone 13 viral titers were measured as described (34). Mice were bled eight days post infection, plasma collected, RNA isolated, and cDNA generated as described above. Q-PCR was performed using LCMV-specific primers Supplemental Table 1. Titers were calculated using a standard dilution series of LCMV stock.
Flow cytometry and cell sorting
Primary murine CD8 T cells were isolated from spleens of mice using a negative selection method with the CD8a+ T Cell Isolation kit II (Miltenyi Biotec) according to the manufacturer’s protocol. Cells were resuspended in 100 μl of FACS buffer (PBS, 2 mM EDTA, and 1% BSA), stained with fluorophore conjugated antibodies for 30 min at room temperature, washed twice with 1 ml FACS, and resuspended in 300 μl FACS buffer. The fluorophore-antibodies used are detailed in Supplemental Table 1: For ex vivo CD8 T cell experiments PD1-Percp-Cy5.5 (RMPI-30), IL6Ra-APC (D7715A7), IL10R-PE (1B1.3a), Rat IgG2b,k -Percp-Cy5.5 (RTK4530), Rat IgG2b,k – APC (RTK4530), Rat IgG1,k – PE (RTK2071) antibodies were used. For LCMV experiments CD8-FITC (2.43), CD62lg-PercpCy5.5 (MEL-14), CD44-AF700 (IM7), PD1-PE (RMPI-30), CD11b-APCCy7 (M1/70), B220-APCC7 (RA3–6B2), F4/80-APCCy7 (BM8), Thy1.1-Pacific Blue (OX-7) and a fixable viability dye (Ghost Dye-v510) were used. For indicated experiments GP33-APC (H-2D9b) LCMV tetramer staining was conducted prior to introducing extracellular epitope antibodies. Visual gating strategy for all analysis is depicted in Supplemental Figure 1: lymphocyte population determined by FSC-A and SSC-A, single cells on FSC-A and FSC-H, viable cells on Ghost Dye v510, removal of non-T cell lineages using a dump gate on CD11b–B220–F4/80–, and finally gating on CD8 expressing cells. Flow cytometry was conducted with a LSRFortessa (BD Biosciences) or sorted with a FACSAria II (BD Biosciences) using BD FACSDiva software v8.0 (BD Biosciences). All data were analyzed, and figures generated using Flowjo v10.6.2.
RESULTS
IL-6 induces STAT3 activation and prolonged elevation of Pdcd1 expression in CD8 T cells
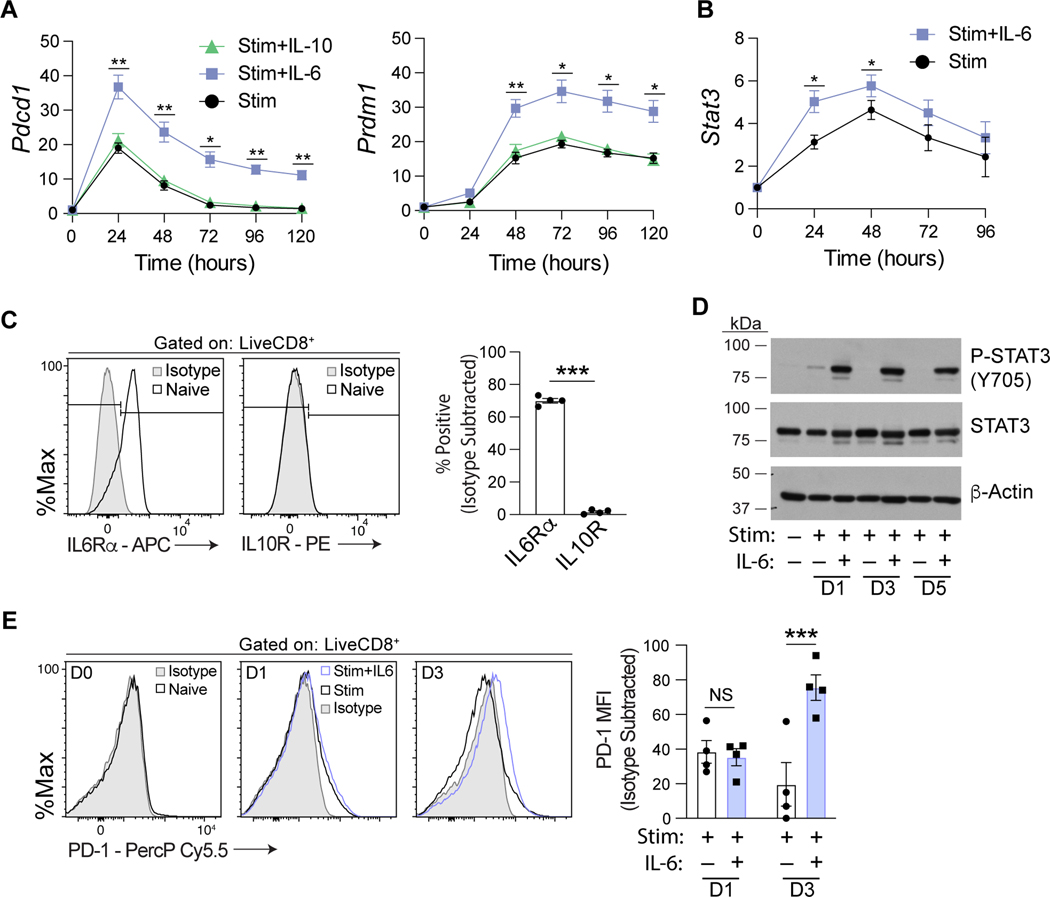
Recent advances in our understanding of Pdcd1 regulation highlight the ability of extracellular cytokine signals, including IL-6, to modulate Pdcd1 expression (20). To determine the temporal kinetics of cytokine treatment on Pdcd1 expression, a time course was conducted on magnetically enriched primary murine CD8 T cells stimulated with anti-CD3/CD28 beads. Isolated CD8 T cells were treated with IL-6 or another STAT3 signaling cytokine, IL-10, for 5 days and Pdcd1 transcript levels measured (Figure 1A). IL-6 treatment led to a significant increase in Pdcd1 mRNA expression over stimulation alone at each point in the time course. Previously it was shown that Pdcd1 mRNA decreases to base line over time due to the induction of BLIMP-1 (26). Intriguingly, the addition of IL-6 to the cultures also increased Prdm1 (encodes BLIMP-1) transcript levels (Figure 1A). Furthermore, IL-6 treatment enhanced Stat3 transcript levels, above stimulation alone (Figure 1B). In contrast, IL-10 treatment did not induce Pdcd1 mRNA expression. To assess IL-6 and IL-10 sensitivity of naïve murine CD8 T cells, surface expression of IL6Rα and IL10R was measured (Figure 1C and Supplemental Figure 1A). Naive CD8 T cells express high levels of IL6Ra supporting the IL-6 specific influence on Pdcd1 and Prdm1 transcription. In contrast, IL10R was not detected above background. As the activities of STAT factors are largely dependent on phosphorylation-marked activation, the phosphorylation status of STAT3 at Y705 after IL-6 treatment was measured (Figure 1D). Indeed, p-STAT3 was dependent on IL-6 and detected over the five days of culture. The observed IL-6 dependent P-STAT3 activation correlated with increased Pdcd1 transcript as well as increased PD-1 surface expression (Figure 1E). Of note, while Pdcd1 transcript increased rapidly after 1 day in the presence of IL-6, an IL-6 specific increase in PD-1 surface levels wasn’t detected until 3 days post IL-6 administration. Taken together, these data implicate IL-6 as a potent and stable driver of Pdcd1, Prdm1, and STAT3 activation.
Figure 1. IL-6 signaling induces prolonged elevation of Pdcd1 transcript and STAT3 activation in CD8 T cells.
(A, B) Splenic CD8 T cells were isolated and cultured ex vivo with anti-CD3/CD28 beads in the presence/absence of the indicated cytokines for up to five days as denoted. Pdcd1, Prdm1, and Stat3 mRNA expression was measured by real-time RT-PCR and the data presented normalized to 18s rRNA levels. Data from 3 independent experiments were averaged ±SEM. (C) Splenic naïve CD8 T cells were magnetically enriched and flow cytometric analysis for IL6Rα and IL10R expression performed. Percent positive for each protein is normalized to an isotype control. Data are representative of four independent experiments and plotted ±SEM. (D) Analysis of splenic CD8 T cells cultured ex vivo as above to assess the phosphorylation status of STAT3 at tyrosine 705 (Y705). Unphosphorylated STAT3 and β-actin levels serve as input controls. Shown is a representative immunoblot of three independent experiments. (E) Splenic naïve T cells were prepared as indicated in A. CD8 T cells were stained with an antibody against PD-1 on Days 0, 1, and 3 following the indicated treatment and flow cytometry was performed. PD-1 median fluorescent intensity measurements were normalized to isotype controls. Data are representative of four independent experiments and plotted ±SEM. Statistical significance was determined by a two-way ANOVA (A, B, and E) or two-tailed t test (C). *, p < 0.05; **, p < 0.01; or ***, p < 0.001.
IL-6 drives STAT3 binding at the Pdcd1 locus in CD8 T cells
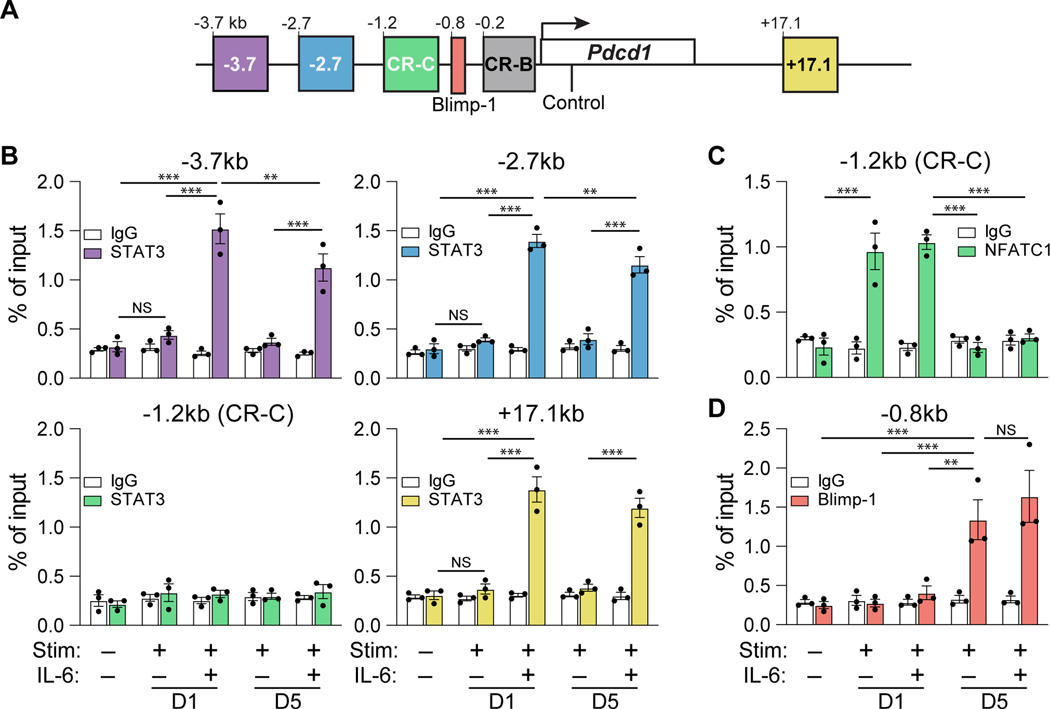
Building on a previous report indicating that STAT3 downstream of IL-6 directly associates with Pdcd1 regulatory regions, a more comprehensive analysis of STAT3 occupation at the Pdcd1 locus was performed (19). Specifically, STAT3 enrichment was analyzed at an expanded number of Pdcd1 regulatory elements, including CR-B, CR-C, −2.7 kb, −3.7 kb, and +17.1 kb (Figure 2A). Such regions were selected through the analysis of previously published DNA hypersensitivity data at the Pdcd1 locus and presence of STAT factor motifs within each region (19, 20, 27). To determine the stability of STAT3 binding at each of these regions, enrichment was determined at both day 1 and day 5 post IL-6 treatment in primary murine CD8 T cells by chromatin immunoprecipitation (ChIP). Consistent with previous findings (19), STAT3 was found to only associate with the Pdcd1 locus in the presence of IL-6, occurring at −3.7, −2.7, and +17.1 (Figure 2B). STAT3 remained significantly enriched at each of these elements five days post IL-6 treatment. CR-C, which binds NFATC1 did not bind STAT3, demonstrating cis-element specificity.
Figure 2. IL-6 drives STAT3 binding at the Pdcd1 locus in CD8 T cells.
(A) Schematic of the Pdcd1 locus detailing the −3.7kb, −2.7kb, and +17.1kb enhancer elements and the CR-C (−1.2kb), CR-B (−0.2kb), and BLIMP-1 binding regions (−0.8kb). (B-D) Primary splenic CD8 T cells were magnetically enriched and subjected to anti-CD3/CD28 stimulation for 1 or 5 days in the presence or absence of IL-6. ChIP employing antibodies against STAT3, NFATC1, and BLIMP-1 were conducted to assess enrichment of each factor within the above sites, with bar color corresponding to specific regulatory regions detailed in (A). Enrichment of non-specific IgG was employed as a negative control for antibody binding. Data are representative of three independent experiments and plotted ±SEM. Statistical significance was determined by two-way ANOVA and denoted as *, p < 0.05; **, p < 0.01; or ***, p < 0.001.
The TCR/NFATC1 signaling axis has been established as a potent inducer of Pdcd1 and is a required prerequisite for IL-6-mediated enhancement of Pdcd1 expression (19). To determine if IL-6 augmented the TCR signaling pathway, occupancy of NFATC1 at CR-C was assessed in the presence of IL-6 (Figure 2C). IL-6 treatment had no effect on NFATC1 binding to CR-C one day post TCR stimulation. Additionally, IL-6 did not prolong the occupancy of NFATC1 at day five, indicating a TCR/NFATC1 independent mechanism of IL-6-specific Pdcd1 enhancement. Interestingly, the observation that Pdcd1 expression is induced despite a similar increase in Prdm1 transcript upon IL-6 treatment (Figure 1A) suggests that IL-6 signaling may be capable of overcoming BLIMP-1 mediated transcriptional silencing. Surprisingly, irrespective of IL-6 treatment, BLIMP-1 was bound to its site within the Pdcd1 locus at day five (Figure 2D). These data suggest that IL-6 dependent STAT3 enrichment at the Pdcd1 locus is dominant to the repressive actions of BLIMP-1.
IL-6 signaling circumvents BLIMP-1 mediated suppression of Pdcd1
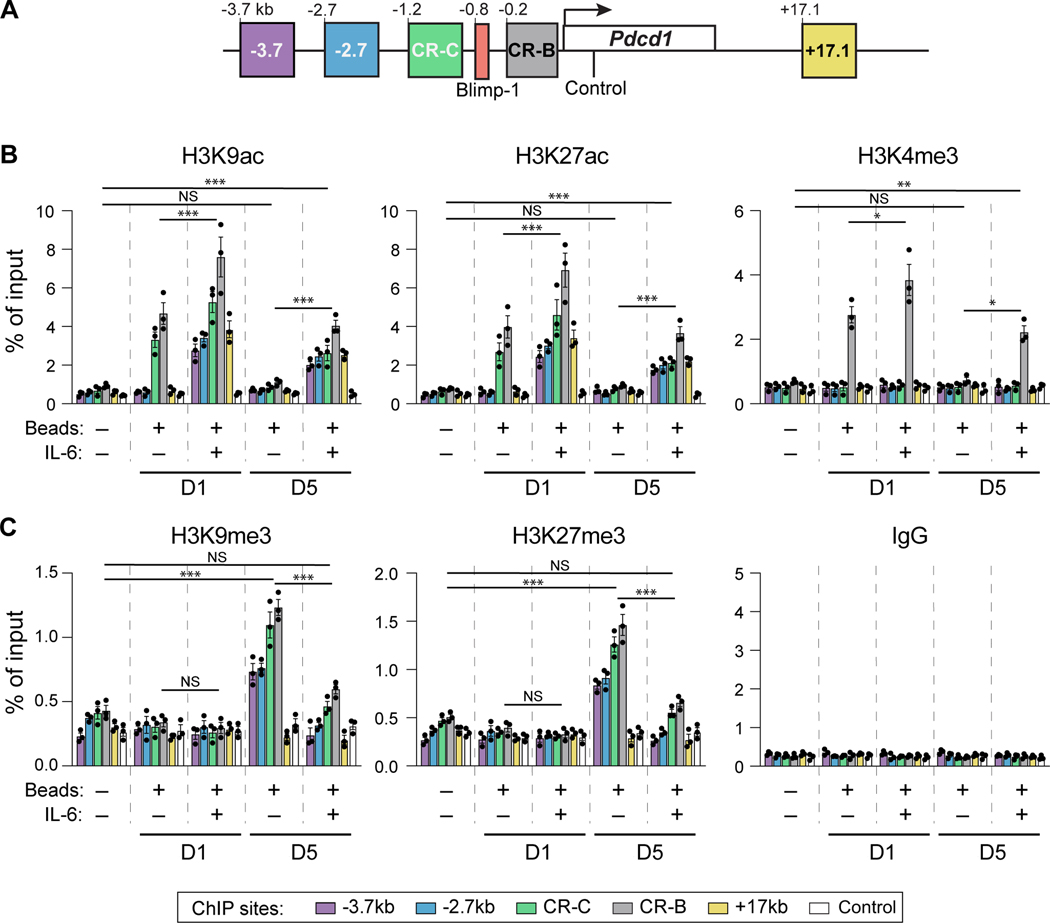
BLIMP-1 is known to promote a repressive epigenetic landscape, in part through influencing the posttranslational histone modifications at key Pdcd1 regulatory regions (26, 27). Thus, one way IL-6 may bypass BLIMP-1 silencing is through influencing the ability of BLIMP-1 to alter the composition of histone modifications at key Pdcd1 regulatory elements. To test this hypothesis, the effect of IL-6 on both active (H3K9ac, H3K27ac), promoter (H3K4me3), and repressive (H3K9me3, H3K27me3) chromatin marks within Pdcd1 regulatory elements were assayed by ChIP (Figure 3A). As BLIMP-1 association with the Pdcd1 locus is delayed, and not detected until day 5 (Figure 2D), histone modifications were assayed at both day one and day five. TCR stimulation induced active histone modifications (H3K9ac, H3K27ac, and H3K4me3, respectively) at day 1 at CR-B and CR-C (Figure 3B). These modifications were lost at day 5, consistent with Pdcd1 mRNA expression presented above. In the presence of IL-6, the above active histone modifications were now detected at day 5 at −3.7, −2.7, CR-C, CR-B and +17.1 and enhanced at CR-B and CR-C compared to stimulation alone. Conversely and consistent with the binding of BLIMP-1, repressive modifications (H3K9me3 and H3K27me3) appear at day 5 but were diminished in their enrichment in cells cultured with IL-6 (Figure 3C). Collectively, these data indicate that IL-6/STAT3 induces Pdcd1 expression, at least in part, by impeding activities attributed to BLIMP-1-dependent epigenetic silencing (26, 27, 35).
Figure 3: Active histone modifications are observed post IL-6 treatment at the Pdcd1 locus.
(A) Schematic of the Pdcd1 locus illustrating the assayed regions used in histone ChIP experiments, including a negative control sequence within the Pdcd1 gene body. (B-C) Primary splenic CD8 T cells were stimulated with anti-CD3/CD28 beads ex vivo and treated with or without IL-6. Cells were harvested after 1 or 5 days and ChIP was performed using antibodies against (B) active histone modifications H3K9ac, H3K27ac, and H3K4me3, and (C) repressive histone marks H3K27me3 and H3K9me3, with IgG enrichment serving as a negative control. Bar color corresponds to the locations within the Pdcd1 locus defined in (A). Data are represented of three independent experiments and plotted ± SEM. Statistical significance was determined by two-way ANOVA: *, p < 0.05, **; p < 0.01; or ***, p < 0.001.
STAT3 is required for IL-6 induction of Pdcd1 in CD8 T cells
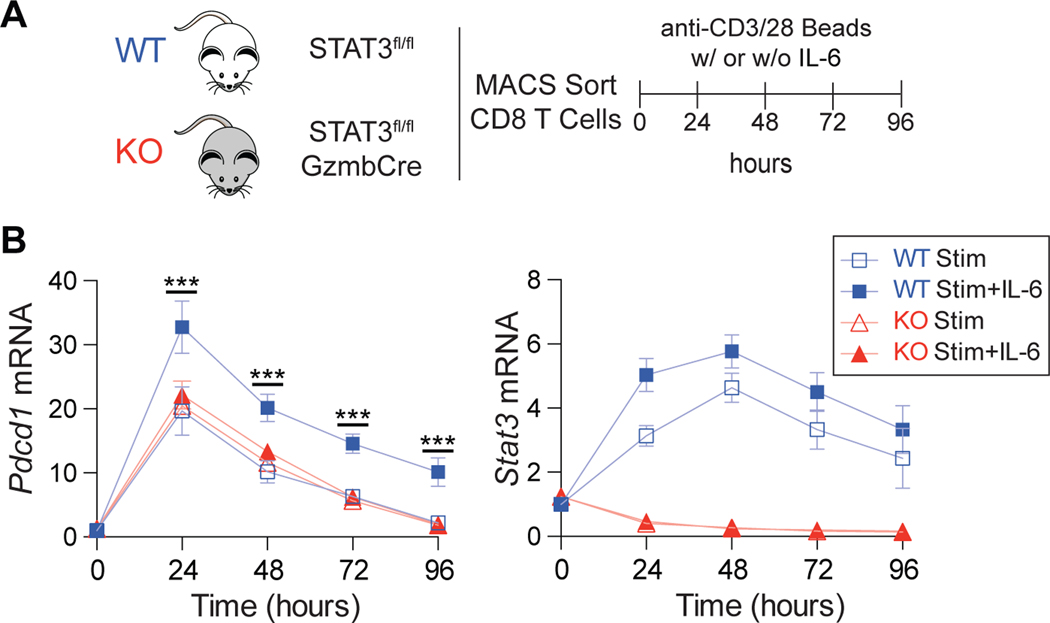
To establish whether STAT3 was essential for IL-6 induction of Pdcd1, Stat3fl/flGzmbCre conditional knockout (KO) mice were bred. This genotype results in Gzmb-dependent expression of Cre recombinase, resulting in a deletion of Stat3 within the activated CD8 T cell compartment (26, 33). Primary naïve CD8 T cells were isolated from both WT and KO mice, stimulated with anti-CD3/CD28 beads with and without IL-6, and cultured for four days (Figure 4A). As above, WT cells treated with IL-6 upregulated Pdcd1; however, STAT3-deficient KO cells failed to enhance and prolong Pdcd1 transcript levels relative to stimulation alone (Figure 4B). As expected, Stat3 transcripts were drastically reduced in the KO mice following anti-CD3/CD28 beads stimulation. These data place STAT3 as a key mediator of IL-6-dependent Pdcd1 induction in vitro.
Figure 4. STAT3 is required for IL-6 induction of Pdcd1 in CD8 T cells.
(A) Schematic of experimental outline. Primary CD8 T cells were isolated from spleens of Stat3fl/flGzmbCRE conditional knockout (KO) mice and WT counterparts and stimulated ex vivo with anti-CD3/CD28 beads in the presence and absence of IL-6 for 4 days. (B) Pdcd1 and Stat3 mRNA expression measured by qRT-PCR every 24 hours exposed to treatment conditions detailed in (A). Data are representative of three independent experiments and plotted ± SEM. Statistical significance was determined by two-way ANOVA: *p < 0.05, **p < 0.01, ***p < 0.001.
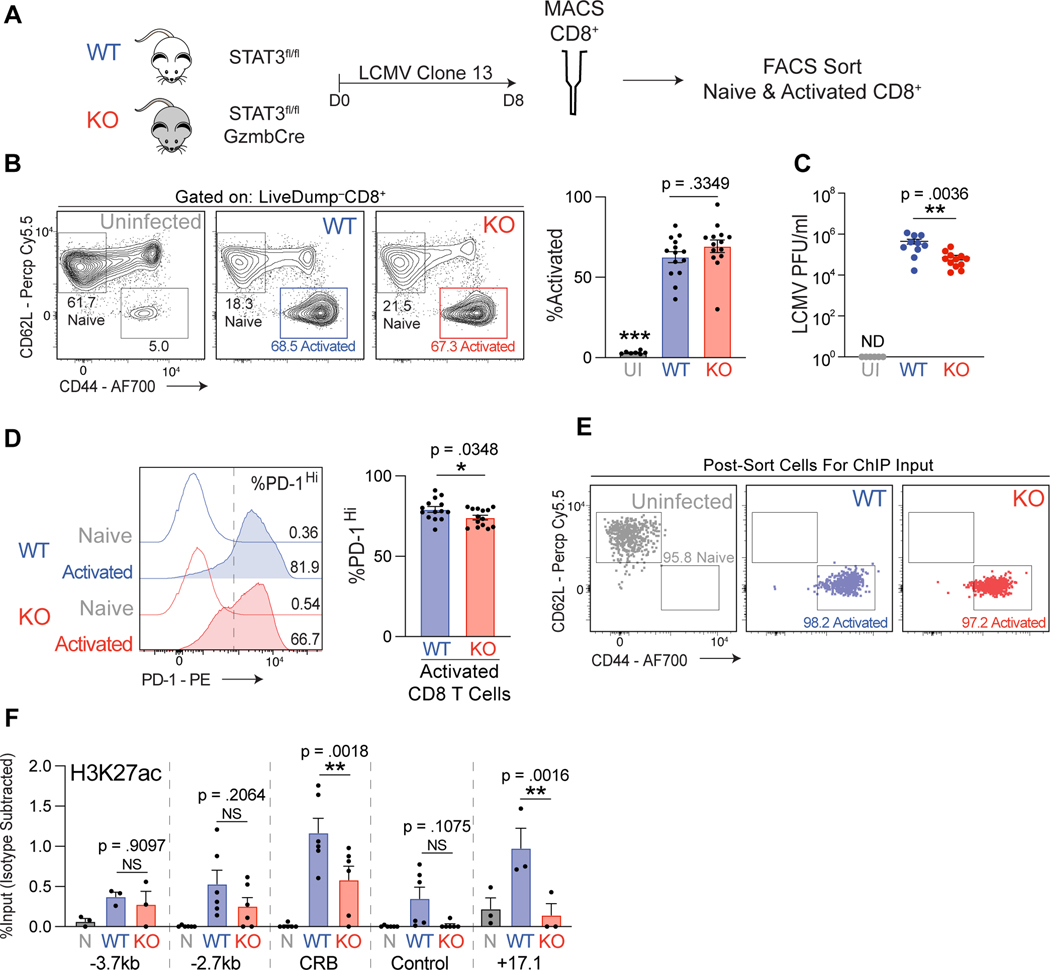
The LCMV clone 13 infection model has been shown to elicit high levels of PD-1 surface protein and Pdcd1 mRNA transcripts in CD8 T cells responding to the infection (7, 36–38). To determine if STAT3 plays a role in governing CD8 immune responses in an in vivo system, WT and KO mice were infected with LCMV clone 13 and after eight days CD8 T cells were magnetically enriched and phenotyped by flow cytometry (Figure 5A and Supplemental Figure 1B). Relative to uninfected controls, the infected WT and KO mice displayed elevated activated CD8 T cell frequencies, although there was no discernable difference in frequency of activated T cells between the two genotypes (Figure 5B). However, significantly lower LCMV virus titers were detected in the peripheral blood of STAT3 KO mice relative to WT mice eight days post infection, suggesting a role for STAT3 in CD8-dependent viral clearance (Figure 5C).
Figure 5. STAT3 is required for IL-6 induction of Pdcd1 in CD8 T cells.
(A) Experimental design for LCMV infection experiments. WT and KO mice were infected with LCMV clone 13. Eight days post infection spleens were collected and CD8 T cells magnetically separated. (B) Frequency of CD44+CD62lg– activated live CD8 T cells in WT and KO spleens post LCMV clone 13 infection determined by flow cytometry. Uninfected WT mice were used as a negative control. (C) LCMV viral RNA was measured from cheek bleeds taken 8 days post infection from WT and KO mice. Plaque forming units/ml (PFU) was calculated with the use of a standard dilution of known concentrations of virus. Uninfected WT mice were used as a negative control. (D) Frequency of PD-1Hi cells in activated CD8 T cells from spleens of WT and KO mice infected with LCMV clone 13. The dotted line on the histogram plot indicates location of gate used to delineate PD-1Hi cells. Data for A-D represent average of four independent experiments with at least 3–4 mice per genotype per infection ±SEM. (E) Flow cytometry dot plots indicating the purity of sorted CD8 T cells as indicated. (F) ChIP data for H3K27ac enrichment at the indicated regions in sorted CD8 T cell populations as indicated. Nonspecific IgG enrichment was subtracted to control for background. Data for E-F represent the average of six independent mice per genotype ±SEM. Statistical significance was determined by two-way ANOVA (B and F) or unpaired two-tailed t test (D-E). *p < 0.05, **p < 0.01, ***p < 0.001.
The observed enhanced viral clearance in the KO mice may be caused in part by impaired PD-1 expression by STAT3 deficient CD8 T cells, as upregulation of PD-1 on CD8 T cells in response to LCMV clone 13 infection is a well-known mechanism to disrupt CD8 T cell functionality (8–13). Intriguingly, LCMV infected KO mice exhibited a modest yet consistently lower frequency of PD-1Hi activated CD8 T cells compared to WT controls (Figure 5D and Supplemental Figure 2). Notably the loss of STAT3 in an in vivo context had a diminished effect on PD-1 expression relative to that observed in an in vitro cell culture environment.
As established in the in vitro system, STAT3 induces PD-1 expression in part through promoting active chromatin marks at the Pdcd1 locus. To determine if the diminished PD-1 expression by STAT3 KO CD8 T cells was accompanied by altered histone modifications, ChIP assays for H3K27ac were performed on sorted activated CD8 T cells from WT and KO mice 8 days post LCMV infection (Figures 5E and 5F). Relative to naïve CD8 T cells isolated from uninfected mice, both WT and KO activated T cell displayed increased H3K27ac throughout the Pdcd1 locus. Correlating with decreased PD-1 expression, KO activated CD8 T cells displayed reduced enrichment for H3K27ac at the CR-B and +17.1 regulatory elements. Collectively, these data depict a role for STAT3 in promoting an epigenetic landscape conducive for heightened PD-1 expression in LCMV-activated CD8 T cell populations.
STAT3 binds to the Pdcd1 locus in LCMV-specific CD8 T cells
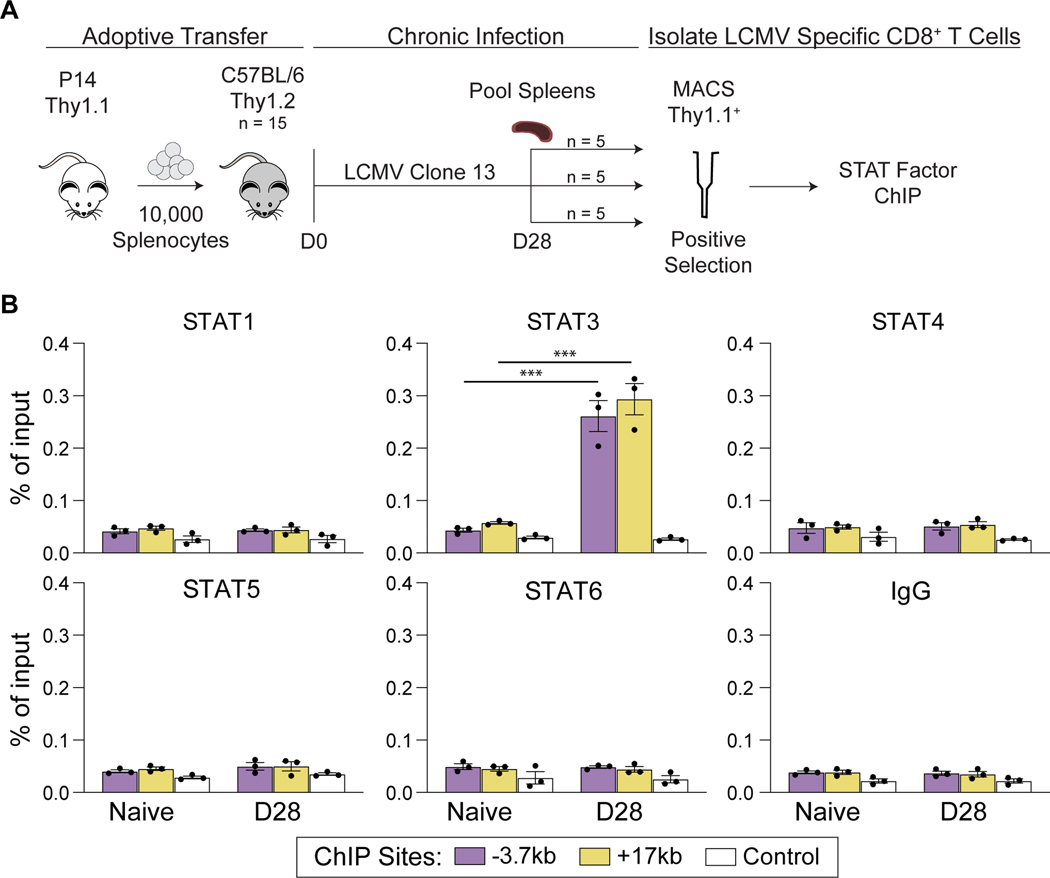
Members of the STAT family are known to have similar binding sequences, often playing redundant roles (39). To gain insight into the specificity and stability of STAT3 in driving Pdcd1 expression in vivo, the enrichment of various STAT factors was assayed in antigen-specific CD8 T cells generated in response to LCMV clone 13 infection (Figure 6A). To achieve robust numbers of antigen specific CD8 T cells, splenocytes from Thy1.1+ P14 mice, which have been genetically engineered to have TCR specific for LCMV clone13 (GP33), were adoptively transferred into Thy1.2+ WT mice. Splenic CD8 T cells were isolated from the recipient mice 28 days post LCMV infection and subsequently enriched for Thy1.1. Flow cytometric analysis of enriched cells revealed a high frequency of Thy1.1 and GP33 antigen-specific cells (Supplemental Figure 1C). Intriguingly, ChIP analysis of the isolated CD8+ Thy1.1+ cells, which can be recovered in high numbers, revealed STAT3 to be significantly enriched at Pdcd1 regulatory regions, while STAT1, 4, 5, and 6 were absent from the locus (Figure 6C). Specifically, STAT3 was found to associate with the −3.7 kb and +17.1 kb regulatory elements, consistent with the in vitro findings (Figure 2B). Thus, STAT3 binds the Pdcd1 locus in antigen-specific CD8 T cells of mice responding to LCMV.
Figure 6: STAT3 binds to the Pdcd1 locus in antigen-specific CD8 T cells.
(A) Outline depicting experimental design. Splenocytes from Thy1.1+ P14 transgenic mice and adoptively transferred into Thy1.2+ WT C57Bl/6 mice, which were subsequently infected with LCMV clone 13. After 28 days, splenic Thy1.1+ CD8 T cells were magnetically isolated and used for STAT factor ChIP. (B) ChIP assays assessing binding of STAT1, STAT3, STAT4, STAT5, and STAT6 at the Pdcd1 locus (as depicted in Figures 2 and 3) post LCMV. IgG was used as a non-specific binding control. Data are presented as % of total input and representative of 3 independent experiments (mean of n = 3 ± SEM). Statistical significance was determined by two-way ANOVA *p < 0.05, **p < 0.01, ***p < 0.001.
DISCUSSION
This study places the IL-6/STAT3 signaling pathway as a regulator of Pdcd1 expression in murine CD8 T cells. Treatment with IL-6 resulted in activation of STAT3 and prolonged and enhanced expression of Pdcd1. Activated STAT3 associated with key Pdcd1 cis-regulatory elements, −3.7, −2.7, and +17.1, yet had no effect on BLIMP-1 association at its binding site within the locus. Moreover, IL-6/STAT3 was able to counter BLIMP-1 mediated epigenetic silencing of Pdcd1 expression, both by promoting/prolonging active histone modifications and preventing/reducing the application of repressive chromatin modifications. Furthermore, CD8 T cells lacking STAT3 displayed diminished Pdcd1 expression in vitro and surface PD-1 expression in an in vivo LCMV clone 13 infection model. This observed loss of PD-1 by STAT3 KO CD8 T cells in vivo was associated with reduced H3K27ac of key Pdcd1 regulatory elements. Importantly, STAT3 was the only STAT family member bound to Pdcd1 cis regulatory elements in antigen-specific CD8 T cells following LCMV clone 13 infection. Together, this study provides mechanistic insight into the observed TCR/IL-6 enhancement of Pdcd1 expression, placing STAT3 as a key molecular regulator of this process.
It is intriguing that Pdcd1 transcripts were observed after one day of IL-6 treatment ex vivo, but protein expression did not appear until day 3, a time point in which untreated cells showed a decrease in surface PD-1 expression. This suggests that the system may be more highly regulated than through the traditional central dogma. Previously, STAT and NFAT factors were shown to bind and associate with the Pdcd1 locus downstream of cytokines and TCR respectively, correlating with increased transcription (19, 21). However, the longevity of these transcription factor-chromatin associations and subsequent mechanisms of transcriptional induction remained unclear. Here, we established that IL-6 signaling resulted in STAT3 enrichment at Pdcd1 enhancer elements (−3.7, −2.7, and +17.1), but had no effect on NFATC1 association at CR-C. Moreover, at an extended 5-day ex vivo time point, NFATC1 binding at CR-C was lost, an event previously shown to be due to BLIMP-1 binding (26), while STAT3 binding remained. This suggests a model wherein initial signals via TCR/NFATC1 result in activation of the locus at CR-C/CR-B (40) followed by a secondary event in the form of IL-6 to exacerbate expression through association of STAT3 to cis-regulatory elements. Indeed, stimulation of CD8 cells was shown to facilitate increased accessibility at the −3.7, −2.7 and +17.1 regions, paving the way for STAT3 and/or other factors to enhance expression (19). In concordance with the transcriptional events, active histone modifications were observed at Pdcd1 regulatory elements; and importantly, in the presence of activated STAT3 the expected repressive modifications attributed to the binding of BLIMP-1 (26, 27, 35) were diminished or prevented from being deposited. Thus, the positive/active role of STAT3 is dominant to BLIMP-1 in controlling the epigenetic landscape of the region.
Insight into how STAT3 may circumvent BLIMP-1 activities is likely derived from its potential interactions with other transcriptional activators, such as ZIPK, Y14, STAP-2, AIOLOS (41–44). Furthermore, and potentially of higher relevance to the observed Pdcd1 induction, STAT3 binding results in chromatin acetylation through its partnership with the histone acetyltransferase P300 and the BRG1 subunit of the SWI/SNF chromatin remodeling complex (45–47). Some studies have indicated that STAT3-dependent gene regulation requires direct recruitment of histone acetyltransferases (48, 49). As such, the detected increase in H3K9ac and H3K27ac at Pdcd1 cis-regulatory elements upon IL-6 treatment here could be a result of P300 recruitment or stabilization of P300 to the locus by STAT3, as well as the recruitment of additional regulatory elements to enhance expression.
While mice lacking STAT3 exhibited a lower frequency of PD1Hi CD8 T cells in response to LCMV infection, the observed reduction was not as robust as seen in ex vivo cultures. Additionally, we noted that the lack of STAT3 did not significantly alter the H3K27ac levels at −3.7 and −2.7kb, despite the fact that we can observe STAT3 binding in ex vivo derived CD8 T cells in response to IL-6 treatment. While this may be due to the in vivo LCMV infection model, or to the fact that low cell numbers were available for the ChIP assay, other explanations are also possible. For example, physiologically, the extracellular cytokine milieu throughout the course of active infection is highly complex, consisting of a plethora of pro-inflammatory cytokines and other signaling molecules with the downstream transcriptional program of CD8 T cells in vivo being the culmination of all present signaling molecules. Thus, other factors may be able to compensate for loss of STAT3 to drive Pdcd1 expression. Of note, other members of the STAT factor family have been shown to bind similar motifs, specifically interferon-gamma-activated sequences, with site-specific co-enrichment of STAT factors having been previously demonstrated (29, 50–52). Although, in the wild-type setting only STAT3 enrichment at Pdcd1 regulatory regions was observed in vivo, it is possible that upon loss of STAT3 other factors may compensate
IL-6/STAT3 signaling has been shown to play a role in the pathogenesis of human and murine viral infections, including LCMV clone 13 infection (53–55). Recent studies have begun to connect elevated serum IL-6 to increased disease severity in SARS-CoV2 infected individuals (56–58). Notably, COVID-19 patients in the intensive care unit (ICU) exhibited elevated serum IL-6 levels relative to a non-ICU control group (59). Moreover, CD8 T cells from the ICU cohort exhibited an exhaustive phenotype characterized by elevated PD-1 expression (59). Intriguingly, therapeutic treatment blocking IL-6/IL6Rα has been used in a subset of severe COVID-19 patients with some success (60). This opens the possibility that perhaps IL-6 in these infections is augmenting PD-1 expression on virus-responding T cells, resulting in reduced activity of these cells.
Collectively, this study provides insight into the regulatory mechanisms and requirements for IL-6/STAT3-dependent Pdcd1 expression in CD8 T cells. Remarkably, initiation of the IL-6/STAT3 pathway functions to counteract BLIMP-1-driven formation of an epigenetically silenced chromatin state within the Pdcd1 locus. Ultimately, acquisition of insight into Pdcd1 regulatory pathways has proven clinical implications, effectively providing the basis for future therapies aimed at manipulating PD-1 expression.
Supplementary Material
Key points:
IL-6 signaling induces STAT3 activation and upregulation of Pdcd1 expression in stimulated CD8 T cells.
STAT3 binds to Pdcd1 cis-regulatory elements and drives a permissive chromatin landscape.
Activated CD8 T cells generated in response to LCMV infection, require STAT3 to fully upregulate PD-1 expression.
ACKNOWLEDGEMENTS
We thank the members of the Boss and Scharer laboratories for scientific input and their careful critiquing of the manuscript, and Royce Butler for orchestrating mouse care.
Footnote:
This work was supported by National Institute of Health grants: RO1 AI113021 to J.M.B, F32 AI161857 to M.D.P.
REFERENCES
- 1.Cerottini JC, Nordin AA, and Brunner KT 1970. Specific in vitro cytotoxicity of thymus-derived lymphocytes sensitized to alloantigens. Nature 228: 1308–1309. [DOI] [PubMed] [Google Scholar]
- 2.Cantor H, and Boyse EA 1975. Functional subclasses of T-lymphocytes bearing different Ly antigens. I. The generation of functionally distinct T-cell subclasses is a differentiative process independent of antigen. J Exp Med 141: 1376–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Golstein P, Wigzell H, Blomgren H, and Svedmyr EA 1972. Cells mediating specific in vitro cytotoxicity. II. Probable autonomy of thymus-processed lymphocytes (T cells) for the killing of allogeneic target cells. J Exp Med 135: 890–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Francisco LM, Sage PT, and Sharpe AH 2010. The PD-1 pathway in tolerance and autoimmunity. Immunol Rev 236: 219–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schietinger A, and Greenberg PD 2014. Tolerance and exhaustion: defining mechanisms of T cell dysfunction. Trends Immunol 35: 51–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arasanz H, Gato-Canas M, Zuazo M, Ibanez-Vea M, Breckpot K, Kochan G, and Escors D. 2017. PD1 signal transduction pathways in T cells. Oncotarget 8: 51936–51945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ahn E, Araki K, Hashimoto M, Li W, Riley JL, Cheung J, Sharpe AH, Freeman GJ, Irving BA, and Ahmed R. 2018. Role of PD-1 during effector CD8 T cell differentiation. Proc Natl Acad Sci U S A 115: 4749–4754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yamamoto R, Nishikori M, Kitawaki T, Sakai T, Hishizawa M, Tashima M, Kondo T, Ohmori K, Kurata M, Hayashi T, and Uchiyama T. 2008. PD-1-PD-1 ligand interaction contributes to immunosuppressive microenvironment of Hodgkin lymphoma. Blood 111: 3220–3224. [DOI] [PubMed] [Google Scholar]
- 9.Ahmadzadeh M, Johnson LA, Heemskerk B, Wunderlich JR, Dudley ME, White DE, and Rosenberg SA 2009. Tumor antigen-specific CD8 T cells infiltrating the tumor express high levels of PD-1 and are functionally impaired. Blood 114: 1537–1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chemnitz JM, Parry RV, Nichols KE, June CH, and Riley JL 2004. SHP-1 and SHP-2 associate with immunoreceptor tyrosine-based switch motif of programmed death 1 upon primary human T cell stimulation, but only receptor ligation prevents T cell activation. J Immunol 173: 945–954. [DOI] [PubMed] [Google Scholar]
- 11.Hui E, Cheung J, Zhu J, Su X, Taylor MJ, Wallweber HA, Sasmal DK, Huang J, Kim JM, Mellman I, and Vale RD 2017. T cell costimulatory receptor CD28 is a primary target for PD-1-mediated inhibition. Science 355: 1428–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barber DL, Wherry EJ, Masopust D, Zhu B, Allison JP, Sharpe AH, Freeman GJ, and Ahmed R. 2006. Restoring function in exhausted CD8 T cells during chronic viral infection. Nature 439: 682–687. [DOI] [PubMed] [Google Scholar]
- 13.Zajac AJ, Blattman JN, Murali-Krishna K, Sourdive DJ, Suresh M, Altman JD, and Ahmed R. 1998. Viral immune evasion due to persistence of activated T cells without effector function. J Exp Med 188: 2205–2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hashimoto M, Kamphorst AO, Im SJ, Kissick HT, Pillai RN, Ramalingam SS, Araki K, and Ahmed R. 2018. CD8 T Cell Exhaustion in Chronic Infection and Cancer: Opportunities for Interventions. Annu Rev Med 69: 301–318. [DOI] [PubMed] [Google Scholar]
- 15.Ishida Y, Agata Y, Shibahara K, and Honjo T. 1992. Induced expression of PD-1, a novel member of the immunoglobulin gene superfamily, upon programmed cell death. EMBO J 11: 3887–3895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zang X. 2018. 2018 Nobel Prize in medicine awarded to cancer immunotherapy: Immune checkpoint blockade - A personal account. Genes Dis 5: 302–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Durgeau A, Virk Y, Corgnac S, and Mami-Chouaib F. 2018. Recent Advances in Targeting CD8 T-Cell Immunity for More Effective Cancer Immunotherapy. Front Immunol 9: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guo L, Wei R, Lin Y, and Kwok HF 2020. Clinical and Recent Patents Applications of PD-1/PD-L1 Targeting Immunotherapy in Cancer Treatment-Current Progress, Strategy, and Future Perspective. Front Immunol 11: 1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Austin JW, Lu P, Majumder P, Ahmed R, and Boss JM 2014. STAT3, STAT4, NFATc1, and CTCF regulate PD-1 through multiple novel regulatory regions in murine T cells. J Immunol 192: 4876–4886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bally AP, Austin JW, and Boss JM 2016. Genetic and Epigenetic Regulation of PD-1 Expression. J Immunol 196: 2431–2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oestreich KJ, Yoon H, Ahmed R, and Boss JM 2008. NFATc1 regulates PD-1 expression upon T cell activation. J Immunol 181: 4832–4839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xiao G, Deng A, Liu H, Ge G, and Liu X. 2012. Activator protein 1 suppresses antitumor T-cell function via the induction of programmed death 1. Proc Natl Acad Sci U S A 109: 15419–15424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Youngblood B, Noto A, Porichis F, Akondy RS, Ndhlovu ZM, Austin JW, Bordi R, Procopio FA, Miura T, Allen TM, Sidney J, Sette A, Walker BD, Ahmed R, Boss JM, Sekaly RP, and Kaufmann DE 2013. Cutting edge: Prolonged exposure to HIV reinforces a poised epigenetic program for PD-1 expression in virus-specific CD8 T cells. J Immunol 191: 540–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Youngblood B, Oestreich KJ, Ha SJ, Duraiswamy J, Akondy RS, West EE, Wei Z, Lu P, Austin JW, Riley JL, Boss JM, and Ahmed R. 2011. Chronic virus infection enforces demethylation of the locus that encodes PD-1 in antigen-specific CD8(+) T cells. Immunity 35: 400–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McPherson RC, Konkel JE, Prendergast CT, Thomson JP, Ottaviano R, Leech MD, Kay O, Zandee SE, Sweenie CH, Wraith DC, Meehan RR, Drake AJ, and Anderton SM 2014. Epigenetic modification of the PD-1 (Pdcd1) promoter in effector CD4(+) T cells tolerized by peptide immunotherapy. Elife 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu P, Youngblood BA, Austin JW, Mohammed AU, Butler R, Ahmed R, and Boss JM 2014. Blimp-1 represses CD8 T cell expression of PD-1 using a feed-forward transcriptional circuit during acute viral infection. J Exp Med 211: 515–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bally APR, Neeld DK, Lu P, Majumder P, Tang Y, Barwick BG, Wang Q, and Boss JM 2020. PD-1 Expression during Acute Infection Is Repressed through an LSD1-Blimp-1 Axis. J Immunol 204: 449–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jin YH, Hou W, Kang HS, Koh CS, and Kim BS 2013. The role of interleukin-6 in the expression of PD-1 and PDL-1 on central nervous system cells following infection with Theiler’s murine encephalomyelitis virus. J Virol 87: 11538–11551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Powell MD, Read KA, Sreekumar BK, Jones DM, and Oestreich KJ 2019. IL-12 signaling drives the differentiation and function of a TH1-derived TFH1-like cell population. Sci Rep 9: 13991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rebe C, Vegran F, Berger H, and Ghiringhelli F. 2013. STAT3 activation: A key factor in tumor immunoescape. JAKSTAT 2: e23010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang Y, van Boxel-Dezaire AH, Cheon H, Yang J, and Stark GR 2013. STAT3 activation in response to IL-6 is prolonged by the binding of IL-6 receptor to EGF receptor. Proc Natl Acad Sci U S A 110: 16975–16980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Heinrich PC, Behrmann I, Haan S, Hermanns HM, Muller-Newen G, and Schaper F. 2003. Principles of interleukin (IL)-6-type cytokine signalling and its regulation. Biochem J 374: 1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ciucci T, Vacchio MS, and Bosselut R. 2017. A STAT3-dependent transcriptional circuitry inhibits cytotoxic gene expression in T cells. Proc Natl Acad Sci U S A 114: 13236–13241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Matloubian M, Somasundaram T, Kolhekar SR, Selvakumar R, and Ahmed R. 1990. Genetic basis of viral persistence: single amino acid change in the viral glycoprotein affects ability of lymphocytic choriomeningitis virus to persist in adult mice. J Exp Med 172: 1043–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shin HM, Kapoor V, Guan T, Kaech SM, Welsh RM, and Berg LJ 2013. Epigenetic modifications induced by Blimp-1 Regulate CD8(+) T cell memory progression during acute virus infection. Immunity 39: 661–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.He R, Hou S, Liu C, Zhang A, Bai Q, Han M, Yang Y, Wei G, Shen T, Yang X, Xu L, Chen X, Hao Y, Wang P, Zhu C, Ou J, Liang H, Ni T, Zhang X, Zhou X, Deng K, Chen Y, Luo Y, Xu J, Qi H, Wu Y, and Ye L. 2016. Follicular CXCR5- expressing CD8(+) T cells curtail chronic viral infection. Nature 537: 412–428. [DOI] [PubMed] [Google Scholar]
- 37.Utzschneider DT, Charmoy M, Chennupati V, Pousse L, Ferreira DP, Calderon-Copete S, Danilo M, Alfei F, Hofmann M, Wieland D, Pradervand S, Thimme R, Zehn D, and Held W. 2016. T Cell Factor 1-Expressing Memory-like CD8(+) T Cells Sustain the Immune Response to Chronic Viral Infections. Immunity 45: 415–427. [DOI] [PubMed] [Google Scholar]
- 38.Im SJ, Hashimoto M, Gerner MY, Lee J, Kissick HT, Burger MC, Shan Q, Hale JS, Lee J, Nasti TH, Sharpe AH, Freeman GJ, Germain RN, Nakaya HI, Xue HH, and Ahmed R. 2016. Defining CD8+ T cells that provide the proliferative burst after PD-1 therapy. Nature 537: 417–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ehret GB, Reichenbach P, Schindler U, Horvath CM, Fritz S, Nabholz M, and Bucher P. 2001. DNA binding specificity of different STAT proteins. Comparison of in vitro specificity with natural target sites. J Biol Chem 276: 6675–6688. [DOI] [PubMed] [Google Scholar]
- 40.Pham LV, Tamayo AT, Li C, Bueso-Ramos C, and Ford RJ 2010. An epigenetic chromatin remodeling role for NFATc1 in transcriptional regulation of growth and survival genes in diffuse large B-cell lymphomas. Blood 116: 3899–3906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sato N, Kawai T, Sugiyama K, Muromoto R, Imoto S, Sekine Y, Ishida M, Akira S, and Matsuda T. 2005. Physical and functional interactions between STAT3 and ZIP kinase. Int Immunol 17: 1543–1552. [DOI] [PubMed] [Google Scholar]
- 42.Ohbayashi N, Taira N, Kawakami S, Togi S, Sato N, Ikeda O, Kamitani S, Muromoto R, Sekine Y, and Matsuda T. 2008. An RNA biding protein, Y14 interacts with and modulates STAT3 activation. Biochem Biophys Res Commun 372: 475–479. [DOI] [PubMed] [Google Scholar]
- 43.Minoguchi M, Minoguchi S, Aki D, Joo A, Yamamoto T, Yumioka T, Matsuda T, and Yoshimura A. 2003. STAP-2/BKS, an adaptor/docking protein, modulates STAT3 activation in acute-phase response through its YXXQ motif. J Biol Chem 278: 11182–11189. [DOI] [PubMed] [Google Scholar]
- 44.Read KA, Powell MD, Baker CE, Sreekumar BK, Ringel-Scaia VM, Bachus H, Martin RE, Cooley ID, Allen IC, Ballesteros-Tato A, and Oestreich KJ 2017. Integrated STAT3 and Ikaros Zinc Finger Transcription Factor Activities Regulate Bcl-6 Expression in CD4(+) Th Cells. J Immunol 199: 2377–2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ni J, Shen Y, Wang Z, Shao DC, Liu J, Kong YL, Fu LJ, Zhou L, Xue H, Huang Y, Zhang W, Yu C, and Lu LM 2014. P300-dependent STAT3 acetylation is necessary for angiotensin II-induced pro-fibrotic responses in renal tubular epithelial cells. Acta Pharmacol Sin 35: 1157–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ni Z, and Bremner R. 2007. Brahma-related gene 1-dependent STAT3 recruitment at IL-6-inducible genes. J Immunol 178: 345–351. [DOI] [PubMed] [Google Scholar]
- 47.Gray MJ, Zhang J, Ellis LM, Semenza GL, Evans DB, Watowich SS, and Gallick GE 2005. HIF-1alpha, STAT3, CBP/p300 and Ref-1/APE are components of a transcriptional complex that regulates Src-dependent hypoxia-induced expression of VEGF in pancreatic and prostate carcinomas. Oncogene 24: 3110–3120. [DOI] [PubMed] [Google Scholar]
- 48.Ray S, Sherman CT, Lu M, and Brasier AR 2002. Angiotensinogen gene expression is dependent on signal transducer and activator of transcription 3-mediated p300/cAMP response element binding protein-binding protein coactivator recruitment and histone acetyltransferase activity. Mol Endocrinol 16: 824–836. [DOI] [PubMed] [Google Scholar]
- 49.Ray S, Boldogh I, and Brasier AR 2005. STAT3 NH2-terminal acetylation is activated by the hepatic acute-phase response and required for IL-6 induction of angiotensinogen. Gastroenterology 129: 1616–1632. [DOI] [PubMed] [Google Scholar]
- 50.Oestreich KJ, Mohn SE, and Weinmann AS 2012. Molecular mechanisms that control the expression and activity of Bcl-6 in TH1 cells to regulate flexibility with a TFH-like gene profile. Nat Immunol 13: 405–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hirahara K, Onodera A, Villarino AV, Bonelli M, Sciume G, Laurence A, Sun HW, Brooks SR, Vahedi G, Shih HY, Gutierrez-Cruz G, Iwata S, Suzuki R, Mikami Y, Okamoto Y, Nakayama T, Holland SM, Hunter CA, Kanno Y, and O’Shea JJ 2015. Asymmetric Action of STAT Transcription Factors Drives Transcriptional Outputs and Cytokine Specificity. Immunity 42: 877–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Villarino AV, Kanno Y, and O’Shea JJ 2017. Mechanisms and consequences of Jak-STAT signaling in the immune system. Nat Immunol 18: 374–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Velazquez-Salinas L, Verdugo-Rodriguez A, Rodriguez LL, and Borca MV 2019. The Role of Interleukin 6 During Viral Infections. Front Microbiol 10: 1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang J, Wang Q, Han T, Li YK, Zhu SL, Ao F, Feng J, Jing MZ, Wang L, Ye LB, and Zhu Y. 2015. Soluble interleukin-6 receptor is elevated during influenza A virus infection and mediates the IL-6 and IL-32 inflammatory cytokine burst. Cell Mol Immunol 12: 633–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Harker JA, Lewis GM, Mack L, and Zuniga EI 2011. Late interleukin-6 escalates T follicular helper cell responses and controls a chronic viral infection. Science 334: 825–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu F, Li L, Xu M, Wu J, Luo D, Zhu Y, Li B, Song X, and Zhou X. 2020. Prognostic value of interleukin-6, C-reactive protein, and procalcitonin in patients with COVID-19. J Clin Virol 127: 104370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Han H, Ma Q, Li C, Liu R, Zhao L, Wang W, Zhang P, Liu X, Gao G, Liu F, Jiang Y, Cheng X, Zhu C, and Xia Y. 2020. Profiling serum cytokines in COVID-19 patients reveals IL-6 and IL-10 are disease severity predictors. Emerg Microbes Infect 9: 1123–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gubernatorova EO, Gorshkova EA, Polinova AI, and Drutskaya MS 2020. IL-6: Relevance for immunopathology of SARS-CoV-2. Cytokine Growth Factor Rev 53: 13–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Diao B, Wang C, Tan Y, Chen X, Liu Y, Ning L, Chen L, Li M, Liu Y, Wang G, Yuan Z, Feng Z, Zhang Y, Wu Y, and Chen Y. 2020. Reduction and Functional Exhaustion of T Cells in Patients With Coronavirus Disease 2019 (COVID-19). Front Immunol 11: 827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rubin EJ, Longo DL, and Baden LR 2021. Interleukin-6 Receptor Inhibition in Covid-19 - Cooling the Inflammatory Soup. N Engl J Med 384: 1564–1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.