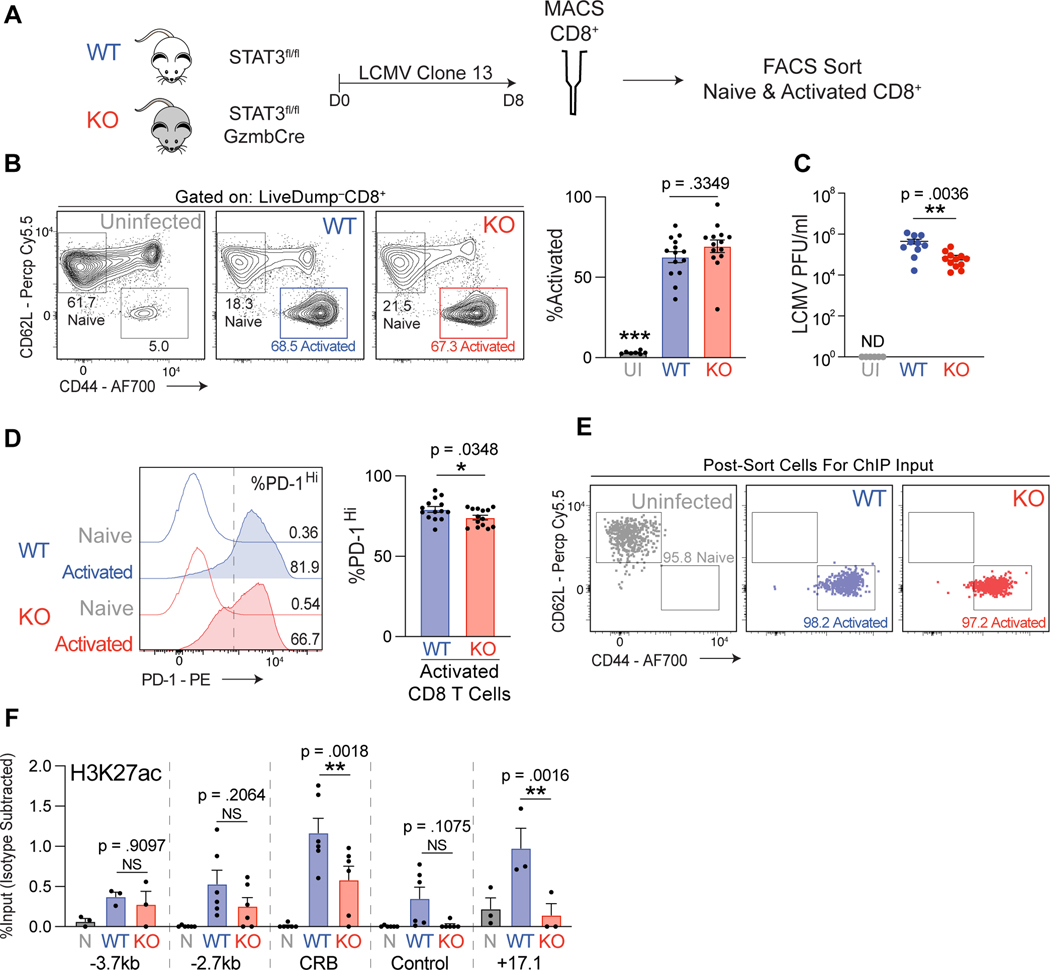
Figure 5. STAT3 is required for IL-6 induction of Pdcd1 in CD8 T cells.
(A) Experimental design for LCMV infection experiments. WT and KO mice were infected with LCMV clone 13. Eight days post infection spleens were collected and CD8 T cells magnetically separated. (B) Frequency of CD44+CD62lg– activated live CD8 T cells in WT and KO spleens post LCMV clone 13 infection determined by flow cytometry. Uninfected WT mice were used as a negative control. (C) LCMV viral RNA was measured from cheek bleeds taken 8 days post infection from WT and KO mice. Plaque forming units/ml (PFU) was calculated with the use of a standard dilution of known concentrations of virus. Uninfected WT mice were used as a negative control. (D) Frequency of PD-1Hi cells in activated CD8 T cells from spleens of WT and KO mice infected with LCMV clone 13. The dotted line on the histogram plot indicates location of gate used to delineate PD-1Hi cells. Data for A-D represent average of four independent experiments with at least 3–4 mice per genotype per infection ±SEM. (E) Flow cytometry dot plots indicating the purity of sorted CD8 T cells as indicated. (F) ChIP data for H3K27ac enrichment at the indicated regions in sorted CD8 T cell populations as indicated. Nonspecific IgG enrichment was subtracted to control for background. Data for E-F represent the average of six independent mice per genotype ±SEM. Statistical significance was determined by two-way ANOVA (B and F) or unpaired two-tailed t test (D-E). *p < 0.05, **p < 0.01, ***p < 0.001.