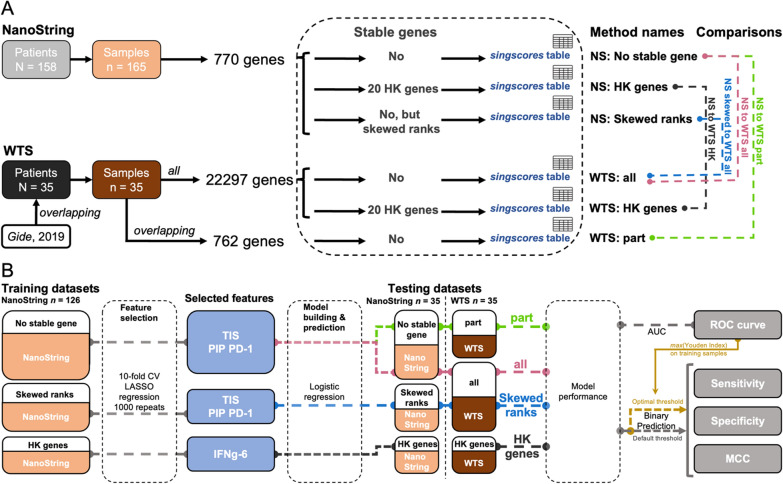
Fig. 1.
Workflow outlining singscores calculation across all samples and cross-platform predictive model building. A The workflow displays several methods to calculate singscores based on different ranking strategies. Both platforms applied 20 genes labelled as HKG in NanoString probes for calibration, named the “HK genes” methods. Without introducing any stable gene, in the NanoString platform singscore directly used such ranks in the “No stable gene” method and used the “Skewed ranks” method based on the regression (Additional file 1: Fig. S7E). The “all” and “part” methods in the WTS platform also did not include any stable gene, but “all” used all genes to rank, and “part” used overlapping genes to rank. “NS” indicates data from the NanoString assay; “WTS” indicates data from the overlapping whole transcriptome Sequencing samples. Four different coloured dot-dashed lines represent four pairs of cross-platform comparisons. B The workflow displays the processes of evaluating multiple cross-platform predictions by signatures’ singscores. The feature selection by tenfold CV LASSO regression and model building based on the three singscore tables derived from three singscore-calculating approaches in A from 126 NanoString samples. Two types of testing datasets were based on 35 overlapping samples from NanoString and WTS platforms. AUC, sensitivity and specificity and MCC were applied to evaluate model performance