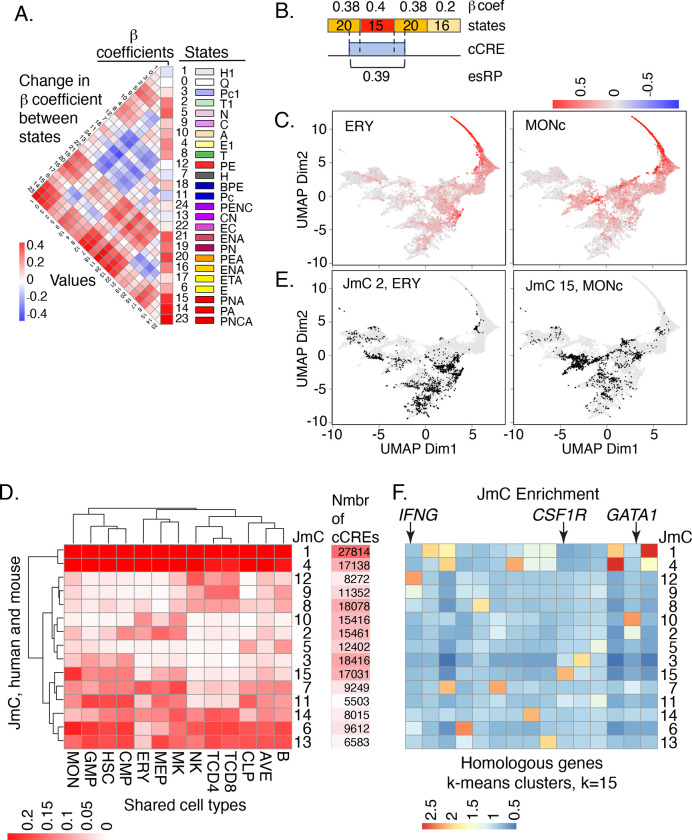
Figure 4. Beta coefficients of states, esRP scores of cCREs, and joint HM metaclusters of cCREs based on esRP scores.
(A) Beta coefficients and the difference of beta coefficients of the 25 epigenetic states. The vertical columns on the right show the beta coefficients, the state ID, state color, and the state names based on the function of all 25 joint epigenetic states. The triangular heatmap shows the difference of the beta coefficients between two states in the right columns. Each value in the triangle heatmap shows the beta coefficient of state on top minus the beta coefficient of the state below based on the order of state in the right columns. (B) An example of calculating esRP score for a cCRE in a cell type based on the beta coefficients of states. For a cCRE covering more than one 200bp bin, the esRP equals the weighted sum of beta coefficients of states that covers the cCRE, where the weights are the region covered by different states. (C) UMAP of cCREs based on their esRP scores across all cell-types. The points are colored by the esRP scores in the designated cell type. (D) The average esRP score of all cCREs in JmC across all cell-types shared by both human and mouse. The right column shows the number of human cCREs in each JmC. (E) UMAP of cCREs based on their esRP scores across all cell-types, with the points colored by the binary label indicating whether a cCRE belongs to the specified JmC. (F) The average enrichment of JmCs in 15 homologous gene clusters. The genes are clustered based on the JmCs’ enrichments by K-means.