Fig. 1.
Study overview and open chromatin broad cell type identification.
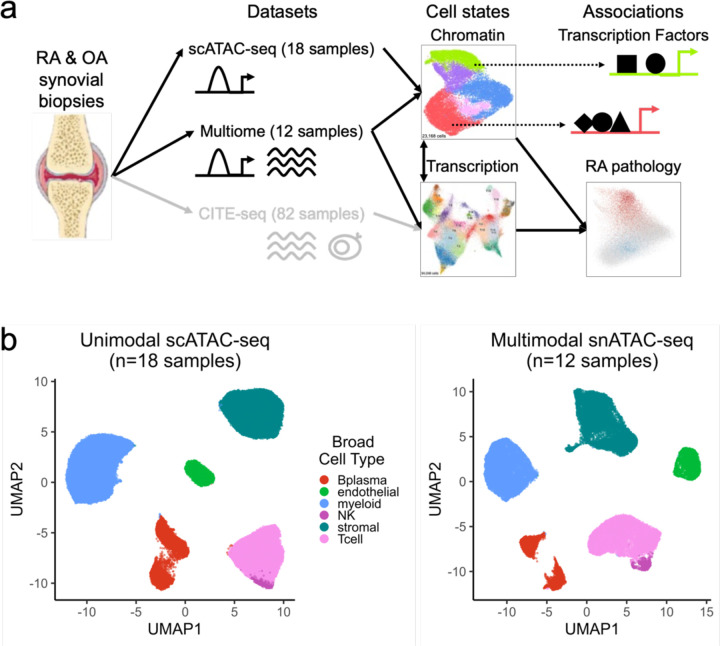
a. Study overview. Synovial biopsies from RA and OA patients were utilized for unimodal scATAC-seq, multimodal snATAC-seq + snRNA-seq experiments. CITE-seq was performed in the AMP-RA reference study12. We defined chromatin classes using the unimodal and multimodal ATAC data and compared them with AMP-RA transcriptional cell states12 classified onto the multiome cells. We further defined transcription factors likely regulating these chromatin classes and found putative links to RA pathology by associating the classes to RA clinical metrics, RA subtypes, and putative RA risk variants.
b. Open chromatin broad cell type identification in unimodal scATAC-seq datasets (left) and multimodal snATAC-seq datasets (right), processed separately.