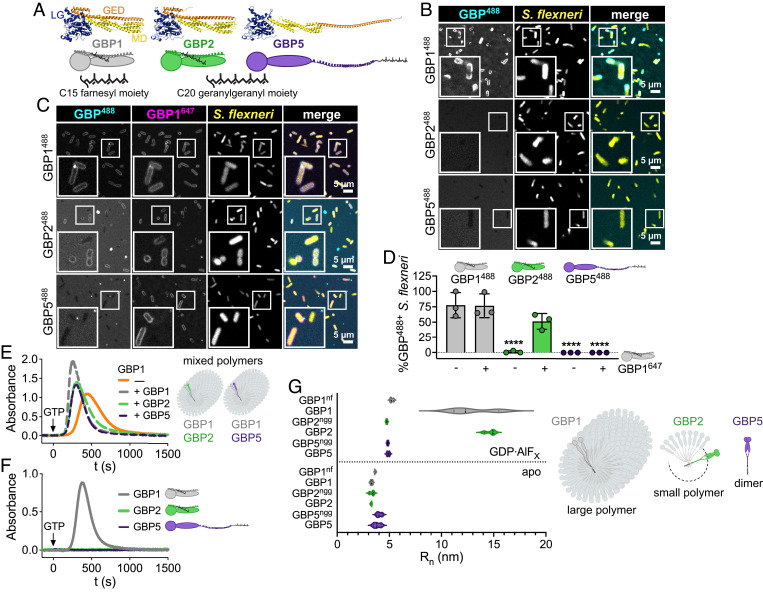
Fig. 4.
GBP2 fails to bind bacteria on its own but forms small polymers. (A) Structures and schematics for GBP1, GBP2, and GBP5. All GBP paralogs consist of a large GTPase domain (LG), a middle domain (MD), and a GTPase effector domain (GED), whereby C-terminal isoprenyl moieties vary with GBP1 becoming post transcriptionally farnesylated, and GBP2 and GBP5 geranylgeranylated. (B–D) Recombinant Alexa Fluor 488-labeled GBP1, GBP2, and GBP5 alone (B) or mixed with Alexa Fluor 647-labeled GBP1 (C) were supplemented with GTP and added to formaldehyde-fixed RFP-expressing S. flexneri. Confocal microscopy time-lapses were recorded, and targeted bacteria were quantified after 1 h (D). (E and F) GBP polymerization was monitored in UV-absorbance-based light scattering experiments. Equal molar ratios of recombinant GBP1, GBP2, or GBP5 were mixed with GBP1, and polymerization was induced with GTP (E). GTP-induced polymerization of single GBPs was monitored over time (F). (G) Number-weighted mean radius (Rn) of nucleotide-free (apo) or GDP·AlFX-bound nonisoprenylated (nonfarnesylated - nf, nongeranylgeranylated - ngg) and isoprenylated GBPs was determined in DLS experiments. (A) PDB entry 1F5N and AlphaFold models AF-P32456, AF-Q96PP8. (B and C) Representative images of three independent experiments. (D and G) Graphs are averages from three independent experiments and are represented by mean ± SD. (D) One-way ANOVA with Dunnett’s multiple comparisons test comparing to GBP1488+ S. flexneri was used, all statistically significant comparisons are shown. ****P < 0.0001. (E and F) Representative graphs from two independent experiments.