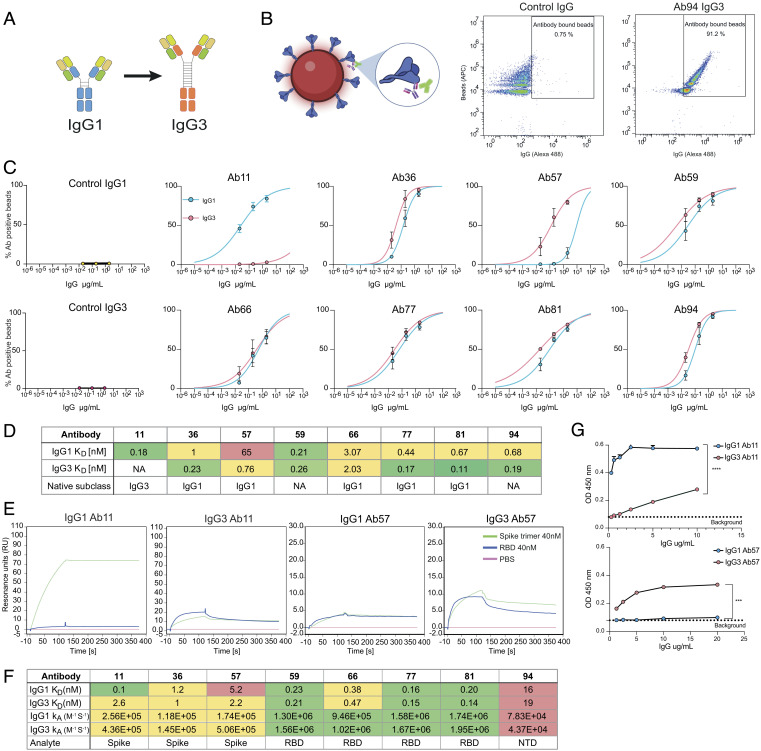
Fig. 1.
Switching the IgG1 constant domain to IgG3 can alter the avidity for Spike protein. (A) Schematic of the heavy and light chain plasmids containing the variable and constant domains. The generation of IgG3 mAbs entails switching the constant domain of the heavy chain from IgG1 (blue) to IgG3 (orange). (B) Spike-coated microspheres are used as a model for SARS-CoV-2 virions. Antibody binding assay is done by opsonizing Spike beads with mAbs and adding a secondary Alexa 488-conjugated antibody that reports IgG binding to Spike beads. (C) Binding curves showing the percentage of IgG positive Spike beads as a function of IgG concentration. Each clone is shown with both subclasses present (IgG1 in blue and IgG3 in red). Three independent experiments were performed with the mean value shown in the graph and error bars representing the SEM. (D) Table summarizing the subclass KD-values and original subclass for each clone. Avidity was calculated by using a nonlinear regression model in GraphPad Prism. (E) Surface plasmon resonance-based binding kinetics with whole spike protein (trimer). Binding of 40 nM Spike-protein (green) to immobilized IgG compared to 40nM RBD (blue) and PBS (pink) for clones 11, 36, and 57 and their respective subclasses. F) Table with KD-values and KA for the different subclasses across all clones (11 to 94) and what analyte (spike, RBD, or NTD) was used to measure it with the SPR-based assay. (G) ELISA data with spike-coated wells and bound IgG is shown for clones 11 and 57 and their respective subclasses. Three independent experiments were performed with titration curves to plot binding curves. Here, one representative experiment with 4 technical repeats is shown with mean value and error bars representing the SEM. Statistical analysis was performed using a two-tailed t test. P value above 0.05 denotes ns, P value below 0.05 denote *, P value below 0.01 denotes **, P value below 0.001 denotes ***, and P value below 0.0001 denotes ****.