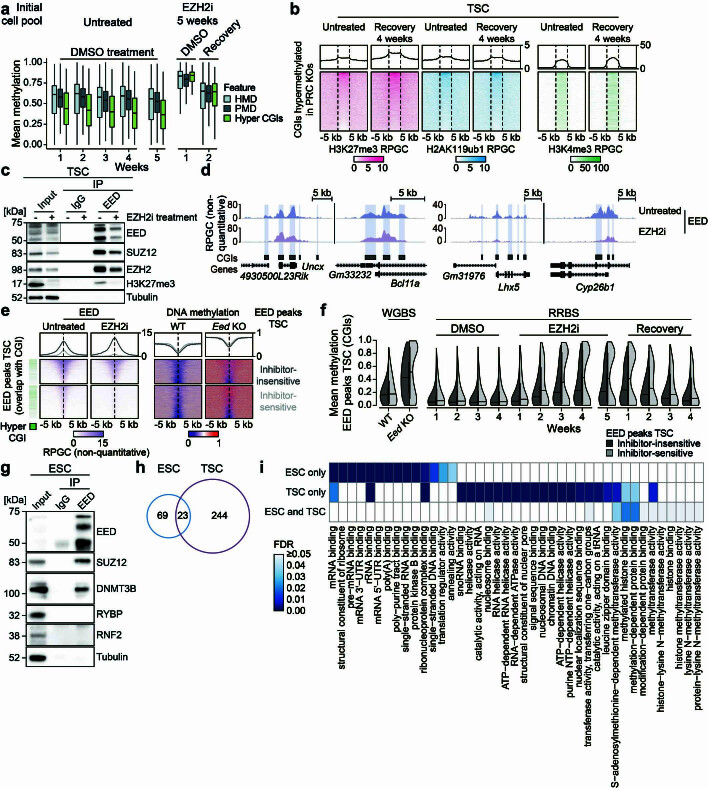
Extended Data Fig. 8. Dynamic interactions between PRC2 and de novo methyltransferases.
a) Feature-level methylation across DMSO controls collected for Fig. 5b (n = 116,056 and 62,434 one kb tiles in HMDs and PMDs, 960 hyper CGIs, single biological replicates). X-axis breaks indicate different experiments (EZH2i treatment and DNMT1i pulse treatment). Lines denote the median, edges the IQR and whiskers either 1.5 × IQR or minima/maxima (if no point exceeded 1.5 × IQR; outliers were omitted). b) MINUTE-ChIP signal heatmaps for EZH2i-recovery and control TSCs (log2 fold change over input). Data is for PRC hypermethylated CGIS (see Extended Data Fig. 6b). H3K27me3 is fully regained after extensive periods of PRC2 inhibition. c) EED Co-IP of EZH2i-treated TSCs. EED is slightly downregulated and preserves interactions with core PRC2 components. Input lanes for the EED blot are taken from the same blot but shown for a higher exposure time given the intensity of the IP lanes. d) Representative genome browser tracks showing EED localization in ~five week EZH2i-treated and control TSCs. Regions with strong EED enrichment maintain signal after EZH2i treatment whereas regions with low enrichment are generally depleted. e) EED signal heatmaps (ChIP-seq) in WT and EZH2i-treated TSCs, centered at EED peaks that overlap CGIs. DNA methylation in WT and Eed KO TSCs are also included. f) Methylation of inhibitor-insensitive and -sensitive EED peaks in WT and Eed KO TSCs (WGBS) as well as for our EZH2i experiments (RRBS, n = 2,868 inhibitor-sensitive and 2,202 -insensitive peaks). White dots denote the median, edges the IQR and whiskers either 1.5 × IQR or minima/maxima (if no point exceeded 1.5 × IQR; minima/maxima are indicated by the violin plot range). g) Co-IP of EED in WT ESCs. EED directly interacts with other components of PRC2 as well as DNMT3B, but not with PRC1 components. h) Overlap of significant EED interaction partners between ESCs and TSCs as determined by IP-MS. i) GO terms for significant EED interaction partners within ESCs and TSCs as determined by IP-MS.