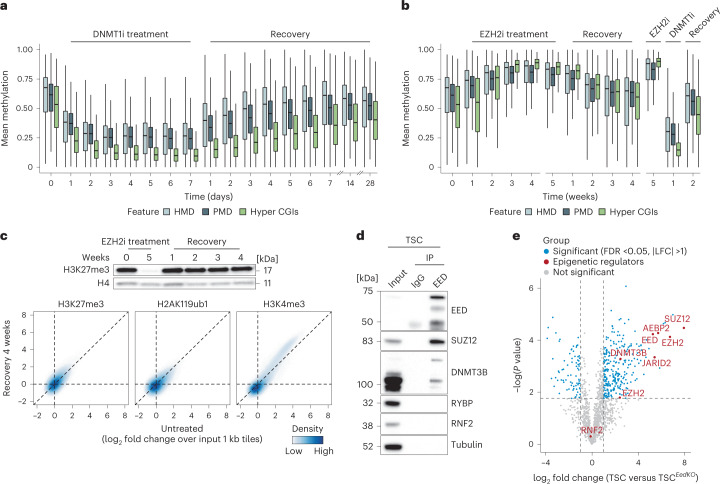
Fig. 5. The TSC epigenome can be reversibly driven to extreme DNA methylation levels.
a, DNMT1i treatment and recovery as measured by RRBS (untreated control measured by WGBS, single biological replicates). Genome-wide methylation drops drastically during the first 3 days and recovers most of the original methylation within one week of withdrawal (n = 117,477 and 62,118 1-kb tiles in HMDs and PMDs, respectively). Hyper CGIs (n = 970) show similar trends, although re-methylation efficiency is slightly lower. Lines denote the median, edges denote the IQR and whiskers denote either 1.5× IQR or minima/maxima (if no point exceeded 1.5× IQR; outliers were omitted). b, EZH2 inhibitor (EZH2i) treatment and recovery time series as measured by RRBS (untreated control measured by WGBS, single biological replicates). Left: genome-wide and CGI methylation rise to extremely high levels after 4 weeks (n = 116,056 and 62,434 1-kb tiles in HMDs and PMDs, respectively, and n = 960 hyper CGIs). The effect is progressively reversed following a 4 week washout period. Right: independent experiment that demonstrates accelerated recovery of steady-state methylation levels by pulse DNMT1i treatment. Lines denote the median, edges denote the IQR and whiskers denote either 1.5× IQR or minima/maxima (if no point exceeded 1.5× IQR; outliers were omitted). c, Top: western blot for H3K27me3 in untreated TSCs, TSCs treated with EZH2i for 5 weeks and weekly recovery timepoints. Bottom: MINUTE-ChIP correlation between untreated and post-recovery TSCs, measured in 1 kb tiles (log2 fold change over input) demonstrate the reversibility of the TSC epigenome. d, Co-IP of EED and core components of PRC1/2, DNMT3B and Tubulin (negative control) in WT TSCs. EED directly interacts with other components of PRC2 as well as DNMT3B, but not with components of PRC1. e, Enrichment and statistical significance of EED interactions within TSCs as measured by MS following IP (WT TSCs were compared with Eed KO to eliminate noise, two-sided Student’s t-test, P values adjusted for multiple testing correction using FDR). EZH2 is plotted twice because of the recovery of two distinguishable isoforms, Q61188;D3Z774 and Q6AXH7, respectively.