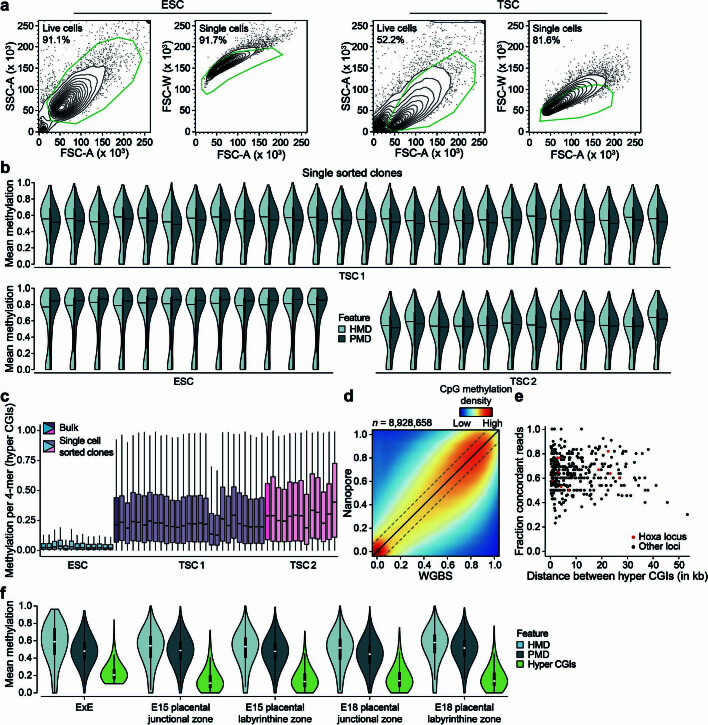
Extended Data Fig. 2. Single cell sorted clones re-gain bulk methylation levels and patterns.
a) Gating strategy for sorting single TSC and ESC clones. FSC-A/SSC-A was used to determine live cells, followed by FSC-A/FSC-W gating for single cells (1 cell/well of a 96 well plate). b) HMD and PMD violin plots (1 kb tiles, n = 99,654 and 51,479 tiles respectively) for single sorted clones. Lines denote the median, edges the IQR and whiskers either 1.5 × IQR or minima/maxima (if no point exceeded 1.5 × IQR; minima/maxima are indicated by the violin plot range). c) 4-mer methylation (n = 21,952) in hypermethylated CGIs for single cell-derived subclones (matching entropy boxplots in Fig. 1d). Subclones from the same cell type have similar methylation levels (low for ESCs, intermediate for TSCs), and resemble in silico generated averages. Lines denote the median, edges the IQR and whiskers either 1.5 × IQR or minima/maxima (if no point exceeded 1.5 × IQR; outliers were omitted). d) CpG-wise comparison of WGBS and Nanopore methylation calls in the same TSC line (single biological replicates, ≥ 10x coverage in both). e) Relationship between CGI pair distance and the fraction of concordantly methylated reads (hypermethylated CGI pairs captured ≥10x). A read is termed concordant if paired CGIs both have methylation levels above or below their unphased averages. Hoxa locus CGI pairs (Fig. 1e) marked in red. The fraction of concordant reads does not appear to depend on distance between CGIs. f) Average HMD, PMD and hyper CGI methylation (n = 274,371, 135,801 and 713 features respectively) values in epiblast, ExE and later placental tissues (E15, E18, two merged biological replicates for epiblast/ExE, six biological replicates for placental tissues). White dots denote the median, edges the IQR and whiskers either 1.5 × IQR or minima/maxima (if no point exceeded 1.5 × IQR; minima/maxima are indicated by the violin plot range). Data from Ref. 34.