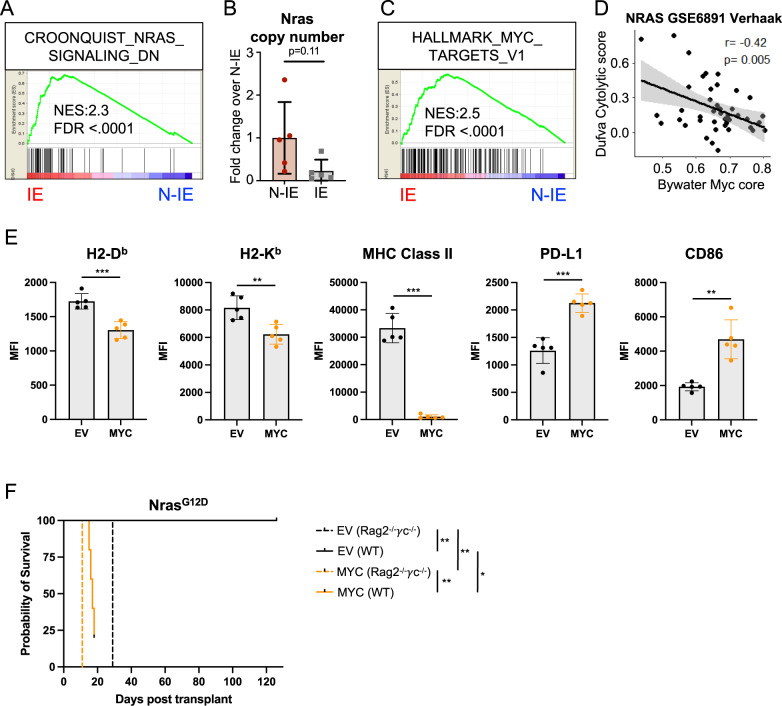
Fig. 5. Myc activation reduces immunogenicity in NrasG12D-driven AML.
A Enrichment of genes correlating with down-regulation of Nras signaling in immunoedited (IE) NrasG12D AML, as determined from RNA-sequencing of GFP+ AML cells isolated from either immunocompetent WT (IE, n = 5) or immunodeficient Rag2−/−γc−/− (N-IE, n = 5) recipients. B Relative quantification of Nras copy number in genomic DNA by qPCR in N-IE (n = 5) and IE (n = 5) NrasG12D AML cells, expressed fold change to the N-IE mean. Each point represents a biologically independent animal, data are presented as mean values +/− SD. C Enrichment of genes correlating with upregulation of MYC transcriptional targets in IE NrasG12D AML. D Correlation between the relative enrichment of a gene set containing core MYC transcriptional targets36 and genes associated with cytolytic infiltrate in AML37, using bulk expression data from NRAS mutant AML patients29. E Flow cytometry analysis of cell surface expression of immune modulatory molecules on N-IE NrasG12D AML cells transduced with either a Myc expression construct (MYC) or empty vector (EV) and passaged in Rag2−/−γc−/− recipients (mCherry+ GFP+ CD11b+ splenocytes from 5 independent Rag2−/−γc−/− recipients) (p = 0.0005 (H2-Db), p = 0.005 (H2-Kb), p = 0.0001 (MHC Class II), p = 0.0002 (PD-L1), p = 0.0079 (CD86)). Data are presented as mean values +/− SD. F Survival of Rag2−/−γc−/− and WT secondary recipients transplanted with 60,000 N-IE NrasG12D/EV or MYC AML cells. Mantel-Cox test for comparison of Kaplan–Meier curves. (n = 5 per group, p = 0.0027 (EV Rag2−/−γc−/− vs EV WT), p = 0.0027 (MYC Rag2−/−γc−/− vs MYC WT), p = 0.0027 (EV Rag2−/−γc−/− vs MYC Rag2−/−γc−/−), p = 0.0133 (EV WT vs MYC WT)) (F). Unpaired two-tailed t-test (E) with Welch’s correction (B). Two-tailed Pearson’s test for association (D). *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. Source data are provided as a Source Data file.